Christoph Bock Lab @ CeMM & MedUni Vienna
@BockLab
Followers
3K
Following
218
Media
169
Statuses
355
Technology-driven biomedical research at CeMM Research Center for Molecular Medicine & MedUni Vienna #cancer #immunology #bioinformatics #AI #singlecell #CRISPR
Vienna, Austria
Joined August 2019
We study cancer and immunology, combining experimental biology with high-throughput technology, bioinformatic methods and machine learning. Our work is interdisciplinary and collaborative. We are equally committed to biological discovery and to advancing medicine.
0
1
32
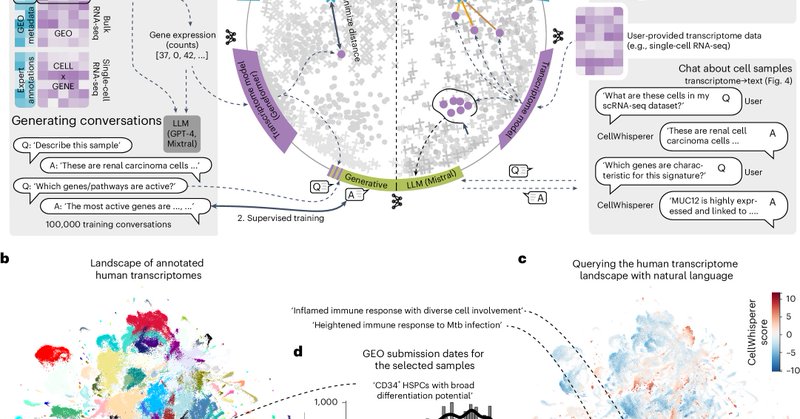
Amazing new tool published @NatureBiotech on interactive analysis of single cell data. The study introduces CellWhisperer, an AI model that links transcriptomes with text descriptions to support natural language queries and zero shot cell type predictions, and integrates it with
4
59
257
CellWhisperer From Transcriptome to Text A great LLM-Chatbot explorer for CELLxGENE & GEO & your own data! Contrastive language image pretraining Geneformer BioBERT >1 million bulk/pseudo-bulk transcriptomes https://t.co/mznOvGrCuj
@muronglizi @PenederPeter @BockLab
0
19
94
Multimodal learning enables chat-based exploration of single-cell data https://t.co/bLy1lvBDYv
3
72
303
🤝 Huge thanks to the team! @muronglizi & @PenederPeter with Daniel Malzl, @salvo_lombardo, @MihaelaPeycheva, Jake Burton, @ahak_hoom, @VarunSh84408069, @TKrausgruber, @celinesin00, @MencheLab, @tomazoulab, @BockLab, @CeMM_News, @MedUni_Wien, @MaxPerutzLabs, @StAnna_CCRI (11/11)
1
1
10
🧬CellWhisperer introduces a chat-based way to explore scRNA-seq data. By enabling natural language analysis, it bridges biologists and bioinformaticians—paving the way for AI-driven bioinformatics assistants. (10/11)
1
0
7
Ready to talk to cells? 🧬 Try the web app with public datasets: https://t.co/QKwGQPAqlm 🖥️ Analyze your own datasets: https://t.co/ms2RE0XtZm 📖 Read the paper: https://t.co/6Suz5VgBc5 (9/11)
nature.com
Nature Biotechnology - CellWhisperer uses multimodal learning of transcriptomes and text to answer questions about single-cell RNA-sequencing data.
1
4
16
📚 We trained on >1 million bulk & pseudo-bulk transcriptomes with textual annotations that we AI-curated from GEO & @CELLxGENE Census. Our training data is open source and useful for developing multimodal biomedical AI models and future bioinformatics research assistants. (8/11)
1
0
8
🪄How does CellWhisperer work behind the scenes? We trained a multimodal AI that links transcriptomes and text, enabling free-text search and annotation of RNA profiles. And we connected this model to an LLM that we fine-tuned into a chat assistant for transcriptome data (7/11)
1
0
7
🚀We also validated CellWhisperer’s chat-based analysis with conventional bioinformatics. CellWhisperer was >4x faster (and 10x cooler 😊). Our recommendation: Use CellWhisperer for dataset exploration – but statistics is still important to ensure rigor & reproducibility (6/11)
1
0
9
🆕 The CellWhisperer paper ( https://t.co/6Suz5VgBc5) includes several new analyses beyond our 2024 bioRxiv preprint ( https://t.co/NwVIX0pqzm). For example, we used CellWhisperer for an AI-guided analysis of human organ development (5/11)
1
1
13
🔬 You can easily query large transcriptome datasets for your favorite biological process using CellWhisperer. Just open Tabula Sapiens ( https://t.co/5YpeJLZv9V) or GEO ( https://t.co/RBou6nt4yt) in CellWhisperer & type your query into the chat box – for example “infection” (4/11)
1
1
14
🔍 We investigate one of the identified cell clusters by selecting the cells & prompting CellWhisperer with ‘Describe these cells in detail’. This interactive workflow is enabled by seamless integration of the CellWhisperer AI chat box into a version of @CELLxGENE Explorer (3/11)
2
0
12
⚙️ To get started, let’s find cells by typing into the CellWhisperer chat box. For example ‘Show me structural cells with immune functions’. CellWhisperer scores each transcriptome by how well it matches this textual query and colors by query match (red: high, blue: low) (2/11)
1
2
14
🗨️ Just published in @NatureBiotech: Our CellWhisperer AI enables chat-based analysis of single-cell sequencing data. You can talk to your cells & figure out the biology without writing any computer code. Paper link and annotated walkthrough in the thread below (1/11)
12
185
689
🗨️Chat with your cells, explore #CellWhisperer! CeMM PI Christoph Bock (@BockLab) together with @MedUni_Wien & @StAnna_CCRI have developed a new #AI tool that lets scientists explore single-cell data using plain English. ➡️ https://t.co/QaFXdfQjyo 📄 https://t.co/DyyIh3FLDO
1
6
11
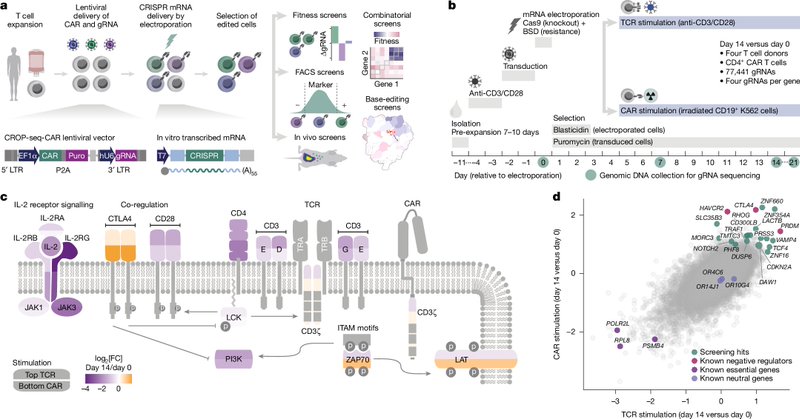
🧬 CAR T cells demonstrate the power of engineered cells as therapeutics. But they fail for most patients. Can we make them better by gene editing? Our paper in @Nature presents a CRISPR platform for optimizing immunotherapies & discovering boosters of CAR T cell function. (1/13)
1
11
20
Vienna researchers tested thousands of gene edits in human CAR T cells and found that deleting RHOG, especially with FAS, makes the cells far more effective against leukemia in mice, offering a new way to boost cancer immunotherapy. https://t.co/joRz5Oe6Yf
nature.com
Nature - CELLFIE, a CRISPR platform for optimizing cell-based immunotherapies, identifies gene knockouts that enhance CAR T cell efficacy using in vitro and in vivo screens.
0
3
12
Elegant ways to rev up the immune system vs cancer keep expanding: Genome editing of engineered T cells https://t.co/5DV0GluxeR
3
39
128
📑Check out our paper titled “Systematic discovery of CRISPR-boosted CAR T cell immunotherapies” at @Nature (open access): https://t.co/LwW4fHrhAf. Feedback & suggestions are very welcome. (13/13)
nature.com
Nature - CELLFIE, a CRISPR platform for optimizing cell-based immunotherapies, identifies gene knockouts that enhance CAR T cell efficacy using in vitro and in vivo screens.
0
1
1
🤝 This was a large project & great teamwork by: P. Datlinger*, E.V. Pankevich*, C.D. Arnold*, N. Pranckevicius, J. Lin, D. Romanovskaia, M. Schäfer, F. Piras, A.-C. Orts, A. Nemc, P. Biesaga, M. Chan, T. Neuwirth, A. Artemov, W. Li, S. Ladstätter, T. Krausgruber, C. Bock (12/13)
1
0
1