Liisa Lutter
@LiisaLutter
Followers
59
Following
161
Media
4
Statuses
34
Postdoctoral researcher in the Eisenberg lab at @UCLA protein folding & misfolding / neurodegeneration / amyloid / structural biology / biophysics
Los Angeles, CA
Joined April 2019
Just in time for Christmas! Excited to see our fibril structures published, showing changing polymorphism of IAPP during the course of amyloid assembly. As always, brilliant team effort from the amyloid team, thanks all @AstburyCentre @naranson
@RadfordLab
https://t.co/iZsk9usR71
2
25
81
Structural reconstruction of individual filaments in Abeta42 fibril populations assembled in vitro reveal rare species that resemble ex vivo amyloid polymorphs from human brains https://t.co/sk7n20u9Wb
#bioRxiv
0
3
5
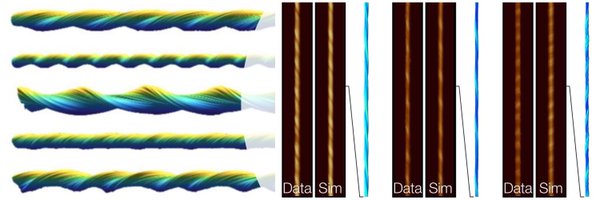
Fantastic to see Trace_y software tools accessible for all to use in their research to do AFM image analysis of helical fibril samples.🤗 https://t.co/0jOfTY3e70
github.com
Contribute to wfxue/Trace_y development by creating an account on GitHub.
Trace_y: Software algorithms for structural analysis of individual helical filaments by three-dimensional contact point reconstruction atomic force microscopy https://t.co/HMRxRbf9kF
#bioRxiv
0
0
3
So proud to hear that Liam won a poster prize at this weeks FASEB meeting on proton aggregation in Dublin for his super exciting data on amyloid fuzzy coats. Well done Liam!!
3
2
37
Check out also this article where @alicepyne discusses why highlighting the research of female scientists in STEM is important, such as closing the gender gap at academic conferences 🙂 https://t.co/MET2iAiWBh
nunano.com
In our blog this month we want to showcase some of the fantastic work being undertaken by women working with AFM today. We asked Dr Alice Pyne to introduce the piece to you and share her thoughts on...
0
0
2
I talked to @NuNanoLtd about my research and experiences as a woman in STEM. https://t.co/tyP9a21CeV
nunano.com
On getting creative as a biochemist user-developer of AFM Dr Liisa Lutter is a postdoctoral researcher in the laboratory of Prof. David Eisenberg at the University of California, Los Angeles (UCLA)....
1
2
16
Our new paper on #tau : Solid-state NMR of paired helical filaments formed by the core tau fragment tau(297-391)
biorxiv.org
Aggregation of the tau protein into fibrillar cross-β aggregates is a hallmark of Alzheimer’s diseases (AD) and many other neurodegenerative tauopathies. Recently, several core structures of patien...
0
3
5
Happy life update 📢: Last week I joined the Eisenberg lab at @UCLA as a postdoctoral researcher. After an incredribly warm welcome I'm excited for new scientific adventures 👩🔬🌿
2
0
7
Integrating individual tau filament AFM 3D envelopes with cryo-EM structures now in @JMolBiol. Demonstrated on an excellent tau297-391 sample by @YoussraHilaly, which was most closely related to PHFs from AD tissue. Illustrative movie below & quantitative comparison in paper.
1
6
15
Happy to share our preprint combining 3 of my favourite things - AFM, cryo-EM & polymorphic amyloid fibrils.🔬 The analysis reveals that unlike heparin-induced tau, dGAE tau fibrils are structurally most closely related to PHFs from #Alzheimer's disease brain tissue.
Structural identification of individual helical amyloid filaments by integration of cryo-electron microscopy-derived maps in ... https://t.co/cgJT0xiNvl
#biorxiv_biophys
0
1
2
Fantastic time meeting everyone in person yesterday ❤️🔬
Wonderful to talk science, amyloid structural biology and cell biology in person again, was a wonderful day with the joint @SerpellLab and @wfxue_lab research away day in East Sussex countryside @Serpell1 💡💡💡
0
0
1
Amazing to be able to contribute with @LiisaLutter @LDAubrey a review of amyloid polymorphism https://t.co/x5Woveg1Gj for @JMolBiol AlphaFold: A Special Issue and a Special time for Protein Science, now out with a fantastic set of 19 great reviews https://t.co/OcOfdJD6wf 😎💡🔬
0
2
4
Today with @emblebi, we're launching the #AlphaFold Protein Structure Database, which offers the most complete and accurate picture of the human proteome, doubling humanity’s accumulated knowledge of high-accuracy human protein structures - for free: https://t.co/vtBGmTkKhy 1/
95
3K
7K
Our recent review on amyloid polymorphism with a twist of AlphaFold 2/structure prediction 😊
My group has recently written a review on our current structural knowledge of the amyloid polymorphism phenomenon. Check out our just-accepted manuscript 💡🔬😎 https://t.co/x5Woveg1Gj
0
0
1
🎉Delighted to share our work out in @Nature today 🎉 using Super-Resolution Localization methods to increase #AFM resolution beyond the limits set by tip radius, reaching at least 4Å resolution. Huge thanks and brilliant working with @ScheuringLab 🙏 https://t.co/bq3djMG0UY
24
247
925
Baker lab's effort at reproducing AlphaFold2 is out on bioRxiv. Pretty impressive performance gains (relative to original trRosetta), if not quite yet at AlphaFold2 level.
biorxiv.org
DeepMind presented remarkably accurate protein structure predictions at the CASP14 conference. We explored network architectures incorporating related ideas and obtained the best performance with a...
4
156
600
Released today @EMDB_EMPIAR & @PDBeurope, & published @CellCellPress, the awesome 3.9 Å #CryoEM structure of the #Flagellar #Motor-Hook complex, the molecular machine that drives rotation of the #Flagellum for #Bacterial motility by Tan et al. from @ZJU_China 1/2
27
461
2K
Been working with @phospho animation on amyloid aggregation. 1st effort is pretty stunning, I think! This is an in vitro, wild-type hIAPP fibril from @R_U_Gallardo @RadfordLab lovely paper in @NatureSMB @Astbury_BSL @AstburyCentre #LoveTheWigglyBits
https://t.co/46omSbeYxd
2
23
124