Jingyou Rao
@JingyouR
Followers
205
Following
732
Media
21
Statuses
179
Incoming Postdoc @UCSF https://t.co/YmbpN6rGip | PhD in Computer Science @UCLA https://t.co/TKqYlWbNIT protein mutational scanning analysis
Los Angeles, CA
Joined July 2018
Our article "Learning antibody sequence constraints from allelic inclusion" is now published in Cell Systems and freely available from this link until Oct 2, 2025: https://t.co/qCZERPUMPF Previous post on the article: https://t.co/ZEiKBWiWKi
Antibodies are highly diverse, but most possible sequences are unstable or polyreactive. In this work led by @milind_jagota, we propose a new source of data for modeling constraints from these properties. Our models show clear improvements in predicting antibody dysfunction.(1/n)
1
10
36
Thanks for posting. I wasn't aware of this beautiful exposition by @rlmcelreath, which I highly recommend for curious statisticians. Primer is now available free of charge: https://t.co/pozbfdiOQW Including Homework and Solution Manual, which we have released for enlightened
@yudapearl @rlmcelreath Did they get to the ending slides? This other video by @rlmcelreath , "Science before Statistics: Causal Inference" is also great: https://t.co/QKGGqC75hd
1
9
47
Try Cosmos in Python now! See vignettes to get started 🔗 Code:
github.com
A Position-Resolution Causal Model for Direct and Indirect Effects in Protein Functions - pimentellab/cosmos
0
0
0
We apply Cosmos to 3 DMS datasets: Kir2.1: abundance → surface expression PDZ3: abundance → CRIPT binding KRAS: abundance → RAF1_RBD binding Cosmos clearly separates direct binding residues from those with indirect effects.
1
0
0
How does it work? Cosmos aggregates mutation effects by position, learns interpretable causal graphs via Bayesian model selection, and outputs residue-level direct vs indirect effects. [5/n]
1
0
0
Cosmos answers three key questions: 1️⃣ Is there a causal link between the phenotypes? 2️⃣ How strong is it? 3️⃣ What would the downstream phenotype look like if we “normalized” the upstream one (i.e., counterfactual inference)? [4/n]
1
0
0
Cosmos is a Bayesian framework that performs residue-level causal inference to decouple how mutations influence upstream vs downstream functions. No need for detailed biophysical models—just stats and data. [3/n]
1
0
0
Multi-phenotype DMS experiments are revealing how mutations impact different protein functions. But these phenotypes are often causally linked—e.g., when measuring activity, we may also capture effects propagated from abundance. So how do we tell what’s direct vs indirect? [2/n]
1
0
0
How do we decouple the effects of two functional phenotypes in protein deep mutational scanning (DMS)? Meet Cosmos, our new statistical framework for causal inference in multi-phenotype DMS. https://t.co/ZOy32xLJ5T [1/n]
biorxiv.org
Multi-phenotype deep mutational scanning (DMS) experiments provide a powerful means to dissect how protein variants affect different layers of molecular function, such as abundance, surface express...
1
0
0
🧬 BREAKING: Our CRISPR-GPT paper is out TODAY in Nature Biomedical Engineering @natBME ! 🤯 We built an AI agent that turns ANYONE into a gene-editing expert in 1 DAY instead of months. An undergrad with ZERO experience achieved 90%+ editing efficiency on their FIRST attempt. 🧵
33
217
742
Presenting at #ISMBECCB2025 tomorrow! GASTON-Mix, a unified model of spatial gradients and domains in spatial 'omics data. 11:20am UK time at RegSys Also recruiting students for my new lab at @JHUCompSci, feel free to reach out if you want to chat
GASTON-Mix: a unified model of spatial gradients and domains using spatial mixture-of-experts https://t.co/N9DTRYq8ns
#biorxiv_bioinfo
1
8
26
Check out our latest foundation model for wearable sensor data (LSM-2)!
Introducing LSM-2, our newest foundation model for wearable sensor data. LSM-2 uses Adaptive & Inherited Masking, a novel self-supervised framework, to learn from incomplete data & achieve strong performance without requiring explicit imputation. More → https://t.co/jeMvzVupZg
0
2
23
🚀 Join CongLab @Stanford! We’re hiring postdocs to create lab-in-the-loop self-evolving AI agents, open benchmarks, to design, test & learn—advancing safer gene & cell therapies. Build on CRISPR-GPT, RNAGenesis model, Genome-Bench, for innovative medicines. #PostdocJobs
1
17
122
Thrilled to share that I just successfully defended my PhD! Thanks to my committee, collaborators, and everyone who’d supported me throughout my seven years at UCLA. A special thank you to my PI Harold for his incredible mentorship! @hjpimentel
4
1
27
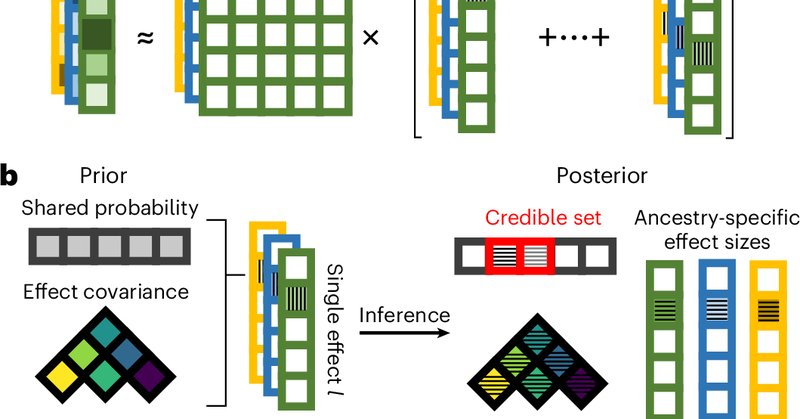
Happy to share that our work from the @nmancuso_ lab is out in @NatureGenet! We developed SuShiE, a multiancestry fine-mapping method for molecular traits.
nature.com
Nature Genetics - SuShiE is a multiancestry fine-mapping method for molecular quantitative trait loci that leverages linkage disequilibrium heterogeneity to improve resolution, infer cross-ancestry...
3
36
98
I’m honored to join Fred Hutch as Professor and Program Head of Biostatistics, and as the Donald and Janet K. Guthrie Endowed Chair in Statistics. Excited to be part of a deeply collaborative and scientifically vibrant community with a rich legacy of impact.
29
14
263
Introducing Arc Institute’s first virtual cell model: STATE
7
110
569
In the nucleus, many intrinsically disordered proteins (IDPs) form condensates. What IDP sequence features drive this behavior? We developed CondenSeq, a high-throughput method to measure nuclear condensate formation & applied it to ~14000 IDPs to find out https://t.co/qfa8jtvFOp
4
38
134
University budgets everywhere are getting slashed, and we hear many PhD students with accepted ICML papers can no longer afford to attend. We are stepping in to offer travel grants for researchers who lost funding. Sponsored by Pillar VC, Ormoni Bio, Latent Labs, and Cimulate
5
35
130
I’ll speak on July 16 (2:30pm ET) at NSF@75: Advancing Statistical Science for a Data‑Driven World @InStatsOrg conference. My talk is about an info-theoretic criterion (ITCA) for combining ambiguous class labels: https://t.co/18yDuDAKwa Free registration:
instats.org
PhD-level seminar on NSF@75: Advancing Statistical Science for a Data‑Driven World (Free Conference) with American Statistical Association . Live Q&A plus on‑demand access. Join via your university’s...
0
5
14