Jason Ernst Lab
@JasonErnstLab
Followers
213
Following
92
Media
1
Statuses
97
A computational biology lab at UCLA
Los Angeles, CA
Joined June 2019
We like to introduce map3C, developed by Joseph Galasso, that drastically improves the mapping and contact calling performance of snm3C-seq and now enables accurate 3D genome modeling. map3C was developed in collaboration with @jernst98 and Frank Alber https://t.co/h8jcF768fR
0
15
42
📣New from Li et al! 📄Whole genome sequence-based association analysis of African American individuals with bipolar disorder and schizophrenia 👉 https://t.co/7wsrScnj3h
0
5
9
GECSI: Large-scale chromatin state imputation from gene expression https://t.co/JDO1iOWu3p
#biorxiv_bioinfo
biorxiv.org
Compendiums of chromatin state annotations based on integrating maps of multiple epigenetic marks such as from ChromHMM have become a powerful resource. While these compendiums have coverage of many...
1
5
9
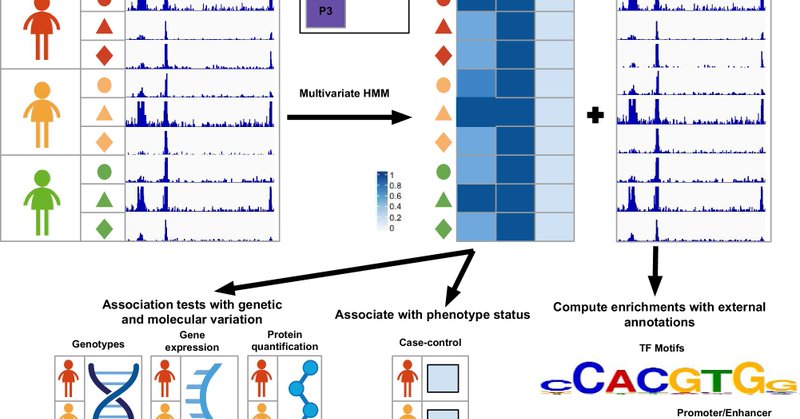
Genome-wide identification and analysis of recurring patterns of epigenetic variation across individuals: Communications Biology, Published online: 07 June 2025; doi:10.1038/s42003-025-08179-5Recurring genome-wide patterns of epigenetic mark variation… https://t.co/GFSqPPuRNA
0
1
2
A new paper: "Genome-wide identification and analysis of recurring patterns of epigenetic variation across individuals" led by Jennifer Zou and @eamaciejewski is now published. https://t.co/R8cIAWtnru
nature.com
Communications Biology - Recurring genome-wide patterns of epigenetic mark variation across individuals learned through computational modeling relate to external measures of molecular variation and...
0
1
1
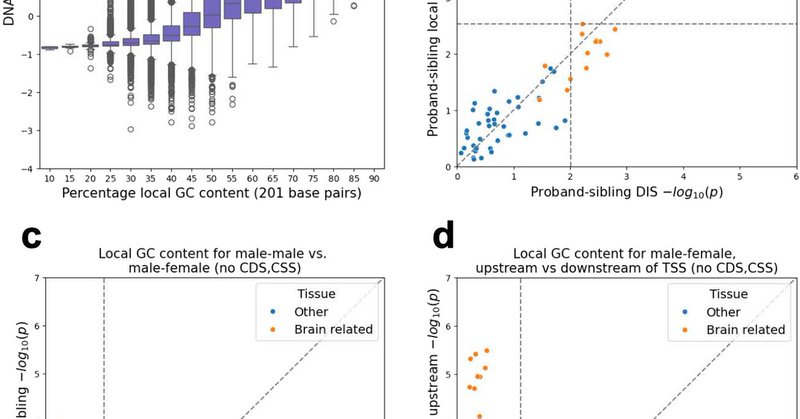
A new paper: "Identifying associations of de novo noncoding variants with autism through integration of gene expression, sequence, and sex information" led by Luke Li is now published
link.springer.com
Genome Biology - Whole-genome sequencing (WGS) data has facilitated genome-wide identification of rare noncoding variants. However, elucidating these variants’ associations with complex...
0
2
2
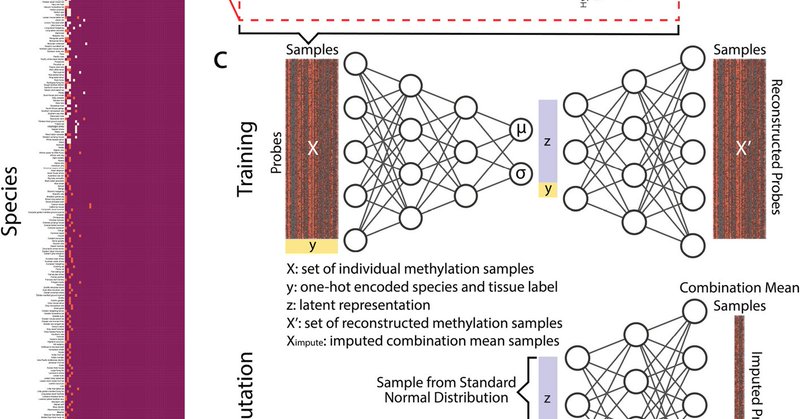
Will comparative biology crack the epigenetic code to longevity? New imputed DNA methylation dataset covers 59 tissues from 348 mammals including long living species such as bowhead whale (211 years), bats, and the naked mole rat. Which tissue holds the key to longevity? Which
pmc.ncbi.nlm.nih.gov
The large-scale application of the mammalian methylation array has substantially expanded the availability of DNA methylation data in mammalian species. However, this data captures only a small...
2
15
60
A new paper "CMImpute: cross-species and tissue imputation of species-level DNA methylation samples across mammalian species" led by @eamaciejewski building on our lab's mammalian methylation array collaboration with @prof_horvath is now published
link.springer.com
Genome Biology - The large-scale application of the mammalian methylation array has substantially expanded the availability of DNA methylation data in mammalian species. However, this data captures...
0
5
9
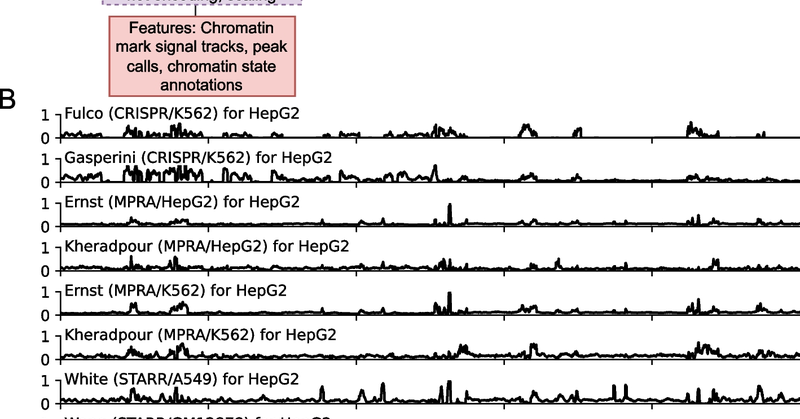
A new paper "ChromActivity: integrative epigenomic and functional characterization assay based annotation of regulatory activity across diverse human cell types" led by @tevzilla is now published.
link.springer.com
Genome Biology - We introduce ChromActivity, a computational framework for predicting and annotating regulatory activity across the genome through integration of multiple epigenomic maps and...
0
3
13
Perspective on recent developments and challenges in regulatory and systems genomics https://t.co/dn8pts7diL is out, amazing to think about this with @JuliaZeitlinger, @ferhatay, @mahonylab, @AMathelier, @AleMedinaRivera, @jernst98!
academic.oup.com
Abstract. Summary: Predicting how genetic variation affects phenotypic outcomes at the organismal, cellular, and molecular levels requires deciphering the
0
13
24
Here is a press release by @UCLAHealth that nicely summarizes the key findings from our paper published today @CellCellPress.
uclahealth.org
1
8
37
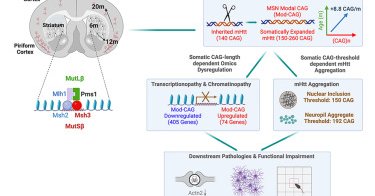
Happy to share our new paper published today @CellCellPress. Thanks to all our coauthors from @dgsomucla, @UCLASemel, and CHDI. Distinct mismatch-repair complex genes set neuronal CAG-repeat expansion rate to drive selective pathogenesis in HD mice: Cell
cell.com
Genes encoding distinct mismatch-repair complexes (i.e., Msh3 and Pms1) set a fast CAG-repeat expansion rate in vulnerable neurons to drive selective and progressive striatal and cortical pathogene...
5
15
77
Learning a Pairwise Epigenomic and Transcription Factor Binding Association Score Across the Human Genome https://t.co/fduik7c4Lb
#biorxiv_bioinfo
0
2
3
Faculty Position in UCLA’s School of Medicine: Broad search in Biomathematics, Biochemistry, Genetics, Pharmacology, Neurobiology, Physiology, Microbiology, Immunology. We seek exceptional scientists committed to research and mentorship. Apply by Jan 19
0
4
4
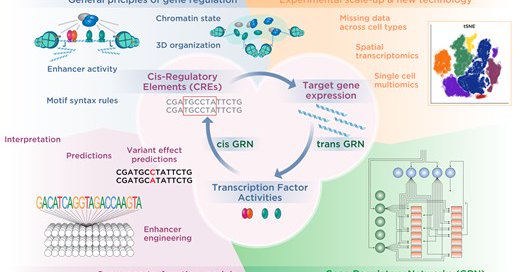
After many zoom meetings, discussions & learning from one another about different ways to look at the same data/problem, our perspective article on regulatory genomics is out. We have a “hallmarks of gene regulation” figure which i am planning to use in my talks. Enjoy reading!
Delighted to have worked on this perspective on the cis-regulatory code with @JuliaZeitlinger, @JasonErnstLab, @ferhatay, @AMathelier, @AleMedinaRivera, @mahonylab, @sinhasaur
https://t.co/fA8KHcp8Sy. We provide an overview of current progress and some challenges to think about.
3
4
26
It was great taking part in writing this perspective on the cis-regulatory code with such excellent scientists. @iscb @ISCB_RegSys has provided us an outstanding platform to share and discuss these topics over the years. Let's keep up the great work in this fantastic community.
Delighted to have worked on this perspective on the cis-regulatory code with @JuliaZeitlinger, @JasonErnstLab, @ferhatay, @AMathelier, @AleMedinaRivera, @mahonylab, @sinhasaur
https://t.co/fA8KHcp8Sy. We provide an overview of current progress and some challenges to think about.
0
2
10
Delighted to have worked on this perspective on the cis-regulatory code with @JuliaZeitlinger, @JasonErnstLab, @ferhatay, @AMathelier, @AleMedinaRivera, @mahonylab, @sinhasaur
https://t.co/fA8KHcp8Sy. We provide an overview of current progress and some challenges to think about.
arxiv.org
Predicting how genetic variation affects phenotypic outcomes at the organismal, cellular, and molecular levels requires deciphering the cis-regulatory code, the sequence rules by which non-coding...
1
24
83
New ‘Aging Clock’ Predicts the Maximum Lifespan of 348 Mammals Including Humans https://t.co/bdaijIFjeY via @Singularity Hub
singularityhub.com
The team says epigenetic signatures linked to gestation, puberty, and maximum lifespan in mammals may be intrinsic to each and difficult to change.
4
31
128
"The maximal lifespan [of people and 347 mammalian species] is strongly associated with an epigenetic signature that is largely independent of sex, body mass, calorie restriction, or other lifestyle factors" @ScienceAdvances
https://t.co/Q3lk3fg4WU
@prof_horvath and colleagues
13
260
827
Identifying associations of de novo noncoding variants with autism through integration of gene expression, sequence and sex information https://t.co/DSQ5PgXTYj
#biorxiv_genomic
0
2
6