Han Kim
@HanKimLab
Followers
532
Following
4K
Media
25
Statuses
1K
Heart, fat, Irx, development and metabolism. Scientist and Associate Professor @HeartInstitute @uOttawa
Ottawa, Ontario
Joined June 2010
Proud of @Yena__Oh, who spearheaded this project, and @RimshahAbid @saif_dababneh @marwanbakr_ @termehaslani for all their contributions. Congrats!
Excited to share our study on the transcriptional regulation of the postnatal cardiac conduction system (CCS) heterogeneity, now out in @NatureComms! https://t.co/xeSotLMFDb
2
0
27
Agents are finally starting to work in biology. We’ve partnered with Anthropic and major biotech vendors - Vizgen, AtlasXOmics, Takara, 10x Genomics - to build a tool that allows scientists to steer their own analysis with natural language. Raw spatial data to publication
26
104
574
#DBfeature Exploring the differentiation potential of EomesPOS mouse trophoblast cells in mid-gestation By Avery McGinnis, Megan Cull, Nichole Peterson, Matthew Tang, Bryony Natale, and David Natale https://t.co/keRI1FZlf2
#Extraembryonic
1
4
27
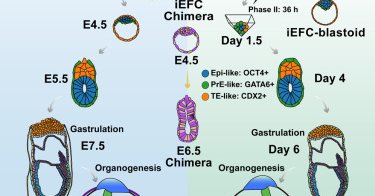
In the latest issue! A complete model of mouse embryogenesis through organogenesis enabled by chemically induced embryo founder cells
cell.com
A small-molecule-only strategy enables reprogramming ESCs into embryo founder cells that give rise to a high-fidelity embryo model directly and efficiently. This embryo model replicates embryogenesis...
1
20
72
Big, beautiful trees!! SMART-PTA for whole-genome+transcriptome on thousand of single cells from the normal human esophagus 🤯 Massively scaling up the power of scWGS to build deep phylogenies and chart somatic evolution from birth throughout life. https://t.co/pciw5yME0x
14
74
243
Please join us WEDNESDAY @ 12PM in the East Conference Room (H-2368) for this week's WIP Rounds. Myriam Adi and Rimshah Abid will present this week. See you there!
0
1
2
Please join us WEDNESDAY @ 12PM in Centre Auditorium for this week's WIP Rounds, a presentation by this year's winner of the Dr. Frans Leenen Trainee Award for Publication Excellence, Dr Yena Oh! For those who attend in person, pizza will be served!
0
3
3
🗞️ "DOLPHIN advances single-cell transcriptomics beyond gene level by leveraging exon and junction reads" ⤵️ @mcgillu @cusm_muhc @Mila_Quebec ▶️ https://t.co/cOZrSgfes2 With #NSERCsupport.
.@mcgillu researchers have developed an artificial intelligence tool that can detect previously invisible disease markers inside single cells. 🧪 https://t.co/wAmNHQ5zhk
@McGillMed @QLSMcGill @NatureComms
0
1
4
My postdoc, Jasper Hof, has made a video tutorial on how to perform a GWAS on the UK Biobank RAP. You can view it at https://t.co/4lNIDFtxKj.
2
45
221
OmniPath ( https://t.co/g2JRIODn2k): integrated knowledgebase for multi-omics analysis https://t.co/SYaCvU76su 🧬🖥️🧪 Python module https://t.co/n7As1pmtmv R package https://t.co/bwzjAmuab8
#Rstats
1
66
306
We’re thrilled to welcome the new UOHI Trainee Committee for 2025-2026! Shout out to the Trainee Committee who made UOHI Summer Student Research Day a success—your passion and discoveries inspire us all! 🔗 Read more: https://t.co/0sarBoRAq8
0
2
6
🚀 Introducing PantheonOS ( https://t.co/NZM3wcHXbG): A Fully Open-Source Agent OS for Science PantheonOS began as a research project in my Stanford lab and has since evolved into a vision to redefine data science in the era of AI—starting with computational biology, especially
2
38
119
UOHI Summer Student Research Day was a resounding success this August 12, 2025! Congratulations to the presentation winners, and we look forward to hosting this event again in 2026!🎉 1st Place Presentation: Richard Wang (Supervised by Dr. Karen Bouchard) 2nd Place
0
3
5
🎉We are pleased to announce the 2025 UOHI Endowed Fellows & Scholars - our next generation of #CVD researchers! Special thanks to @HeartFDN @HeartInstitute & @uOttawaMed Cardiac Endowment Fund at HI for their generous support. #LoveOttawaResearch 🔗 https://t.co/wn3cdthxsp
0
4
5
Congratulations @marwanbakr_ !!
🎉Congratulations to our #BCVS25 Early Career Poster Competition winners! Keep up the amazing science! @AHAScience @BarefieldD @ChiFungLeePhD @S_HamiltonLab @peihengg_heart @SusmitaSahooPhD @DrHelenECollin1 @psatolab @mingtao_zhao @mkontari @SeanM_Wu @arianneinthelab @JonPHeart
0
0
4
A conserved coupling of transcriptional ON and OFF periods underlies bursting dynamics | Nature Structural & Molecular Biology https://t.co/x7YIJQduDu
nature.com
Nature Structural & Molecular Biology - Gene activity levels predict how often and how strongly genes turn on and off. Using live imaging in fruit fly embryos, the authors uncover a conserved...
2
18
89
Happy to share our recent publication on benchmarking peak calling methods for CUT&RUN https://t.co/WVvWyMFGyG
@OxfordJournals Bioinformatics | @AminNoorani In this work, we compared four popular methods - MACS2, SEACR, GoPeaks, and LanceOtron, in a subset of @4DNucleome
1
10
65
Our preprint describing the Range Extender element, which is required and sufficient for long-range enhancer activation at the Shh locus, is out in @Nature. https://t.co/qFrjw55BtX
How do enhancers work over distances that sometimes exceed megabases? Excited to share our work led by @gracecbower where we uncover a unique sequence signature globally associated with long-range enhancer-promoter interactions in developing limb buds: https://t.co/n0p7BdxSdq 1/
18
56
249
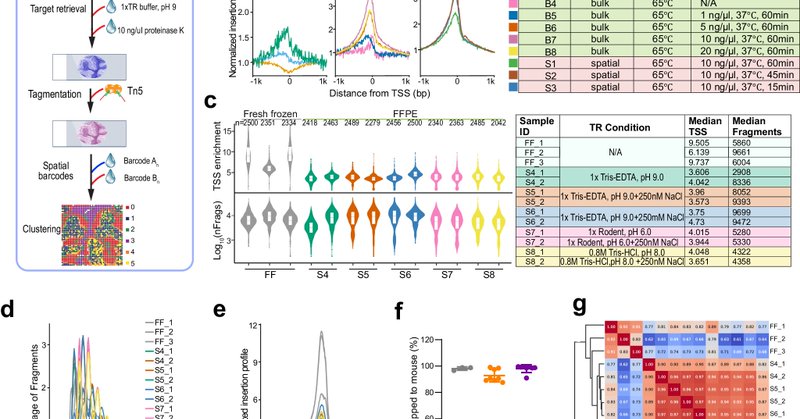
Our spatial FFPE-ATAC-seq is now published in @NatureComms! With extensive optimizations, it performs well across diverse tissues, unlocking the vast FFPE archive for spatial epigenomics. Big congrats to Pengfei Guo @pengfeo, Yufan Chen, and the team!
nature.com
Nature Communications - FFPE tissues are widely used to preserve clinical specimens, but formalin-induced crosslinking limits their use in epigenomic profiling. Here, the authors present spatial...
10
53
233
AI in medicine: what happened this week? 🧵 1. AlphaGenome It's an AI model for understanding DNA It could be used to discover new biology, uncover the clinical impact of mutations, find new drug targets – and help us build a virtual cell for in silico experiments
3
22
143
STAMP it OUT: Single cell Transcriptomics Analysis & Multimodal Profiling- cheap profiling of MILLIONS of cells, multimodal, pertubation expts & tissue atlas'ing at scale. Thanks to brilliant colleagues @LGMartelotto @hoheyn & our amazing teams. Paper:
cell.com
Single-cell transcriptomics analysis and multimodal profiling (STAMP) by imaging enables single-cell analysis of cells in suspension without the need for sequencing. The markedly reduced costs and...
St. Jude researchers and international collaborators developed a more affordable and accessible technique of single-cell RNA analysis that combines microscopy with RNA detection. https://t.co/JKStTlJ6YX
0
6
18