Bin Chen
@DrBinChen
Followers
644
Following
2K
Media
26
Statuses
642
interested in big data + AI + drug discovery. PI @MSU, @UCSF. Trained @ IU, Stanford, Merck, Pfizer and Novartis. 来自重庆土家山寨.
United States
Joined May 2009
RT @jcheminf: We are looking for 2 new Associate Editors who share the vision for our journal to support the Editors-in-Chief in handling….
jcheminf.biomedcentral.com
Journal of Cheminformatics is a fully open access journal publishing original peer-reviewed research in all aspects of cheminformatics. Coverage includes, but ...
0
3
0
I shared some of my memories of my former mentor @atulbutte who has passed away.
linkedin.com
Seven years ago, when I left Atul to begin my independent career, I quoted an old Chinese saying at my farewell party: “A teacher for a day, a father for life.” As this year’s Father’s Day approach...
0
0
5
RT @MSUMD: Congratulations to Teresa K. Woodruff, PhD, on receiving the National Medal of Science! Dr. Woodruff is the first MSU faculty me….
msutoday.msu.edu
MSU Research Foundation Professor Teresa Woodruff, a global leader in fertility science and creator of the oncofertility field, receives the National Medal of Science. Recognized for her groundbrea...
0
2
0
RT @aaguirrecano: The MSU Pharmacology & Toxicology has just posted a tenure-system Associate/Full Professor position for a visionary MSU D….
0
1
0
RT @ruairirobertson: A new bank of >2000 scientific/medical images and icons for free use in figures and presentations just released by @NI….
0
53
0
a great lineup of speakers. In silico Drug Discovery Workshop starting now
ncats.corsizio.com
Due to unprecedented interest in this workshop, we are no longer accepting registrations. However you may view this workshop live on the NIH VideoCast site: Day 1: Wednesday, October 23, 2024, 11:00...
0
0
2
RT @labs_mann: Can a deadly rash be cured by repurposing drugs? Our @Nature paper says yes! Deep Visual Proteomics reveals JAK inhibitors a….
0
23
0
RT @TheMarkFdn: Open RFP: The Mark Foundation's Drug Discovery Award program is accepting letters of intent! This grant offers up to $1M in….
0
10
0
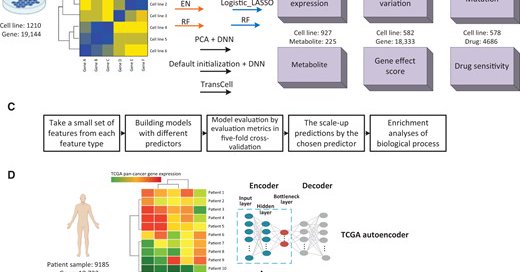
TransCell: In Silico Characterization of Genomic Landscape and Cellular Responses by Deep Transfer Learning . It took one year to formally publish it after acceptance.
academic.oup.com
Abstract. Gene expression profiling of new or modified cell lines becomes routine today; however, obtaining comprehensive molecular characterization and ce
0
0
1
RT @DrBinChen: 1/ Happy to share SPIDER is out @CellSystemsCP. Imputing abundance of over 2,500 surface proteins from single-cell transcri….
0
4
0
Thank you @NICHD_NIH for supporting two of our outstanding postdocs @michiganstateu. Proud to serve as the mentor for @rama_shankar12 's #K99 and as a co-mentor for Dr. Paul's #K99. This means a lot to them and me.
1
0
8
This project was led by my PhD student, Ruoqiao Chen, with significant contributions from Jiayu Zhou on model development. For those interested in SPIDER, please check out our R package (. We welcome your feedback!.
github.com
Contribute to Bin-Chen-Lab/spider development by creating an account on GitHub.
0
0
1
4/ This work was also inspired by another project (, where we demonstrated that it is feasible to predict protein expression (and other genomic features) from gene expression.
academic.oup.com
Abstract. Gene expression profiling of new or modified cell lines becomes routine today; however, obtaining comprehensive molecular characterization and ce
1
0
1
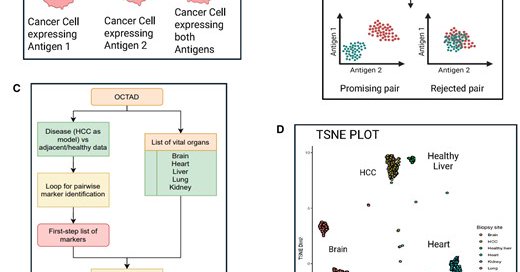
3/ This work was motivated by our recent project (, from which we realized that most antibody-based therapies target surface proteins and it is crucial to consider protein expression in addition to transcript expression during target identification.
academic.oup.com
AbstractMotivation. Bispecific antibodies (bsAbs) that bind to two distinct surface antigens on cancer cells are emerging as an appealing therapeutic strat
1
0
1
1/ Happy to share SPIDER is out @CellSystemsCP. Imputing abundance of over 2,500 surface proteins from single-cell transcriptomes with context-agnostic zero-shot deep ensembles
1
4
5