CVR Bioinformatics
@CVRbioinfo
Followers
2K
Following
3K
Media
129
Statuses
2K
Welcome to our Twitter feed. We’re a group of computer-based researchers, part of the MRC-University of Glasgow Centre for Virus Research (CVR) @CVRinfo
Glasgow, Scotland
Joined April 2016
As the collaborating centre of @WOAH, we’re thrilled to launch our 8th annual course on viral bioinformatics & genomics! Welcome speech by @CVRinfo director @emcat1 & head of Bioinformatics @robertson_lab 🧬📊 #bioinformatics #genomics
1
12
32
Really pleased to share this study from researchers at @CVRinfo and ERASMUS MC, as well as collaborators from RVC, the Pirbright Institute and CRUK Scotland Institute:
nature.com
Nature Communications - H5N1 avian influenza viruses caused an outbreak in dairy cattle. We show that the potential for avian viruses to replicate in cow cells varies across H5N1 evolution,...
1
5
6
Interested in our research in Malawi? Have a listen to @CVRinfo Clinical Viral Epi Group student, @Mhairi_Jan’s podcast on her work understanding #SARSCoV2 immunity in urban & rural Malawi during the #COVID19 pandemic. @MEIRU_MALAWI @MalawiHealth @Animal_Viruses 🦠🧫🩸📈🇲🇼🎙️
🎙️ Mini Podcast | Why Immunity Looks Different Across Malawi's Communities New podcast with @Mhairi_Jan explores new research tracking COVID-19 immunity in Malawi. https://t.co/3OJ6Q5tGef
0
4
6
📰 New CVR research shows that adaptation of bird flu viruses to dairy cattle is driven by changes to the virus over time. This means new variants of bird flu could also infect cows and potentially other mammals in the future. 👉Read more here: https://t.co/fa5M5QegT8
1
7
8
0
3
9
0
3
9
Feeling festive? Why not deck the halls with @CVRinfo's papercraft #VirusSnowflakes and celebrate the science of viruses and the fight against viral diseases. Download for free: https://t.co/Q8Z4L9Dzo0
#scicomm #sciart #decorations
1
8
30
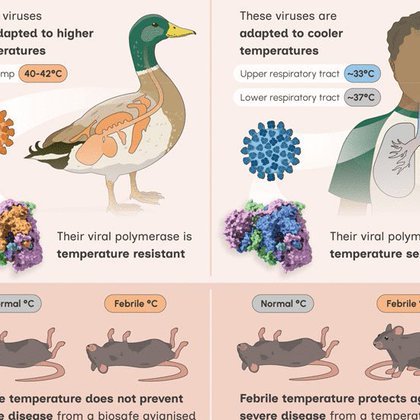
📃 New research, led by CVR & @Cambridge_Uni, shows bird flu viruses can withstand fever temperatures, making them a serious threat to humans. Scientists identified a key gene that explains this resilience, helping us better understand outbreak risks. https://t.co/po6kZi5dug
1
12
14
Really pleased to share this study about the role of fever in fighting influenza A viruses from @WilsonLabCITIID @CVRinfo @MedCambridge :
science.org
Host body temperature can define a virus’s replicative profile—influenza A viruses (IAVs) adapted to 40° to 42°C in birds are less temperature sensitive in vitro compared with human isolates adapted...
2
5
5
Ad hoc, post hoc and intrinsic-hoc in bioinformatics https://t.co/B9SETFduBP (read free: https://t.co/WLvQmp7vXB) 🧬🖥️🧪
0
2
9
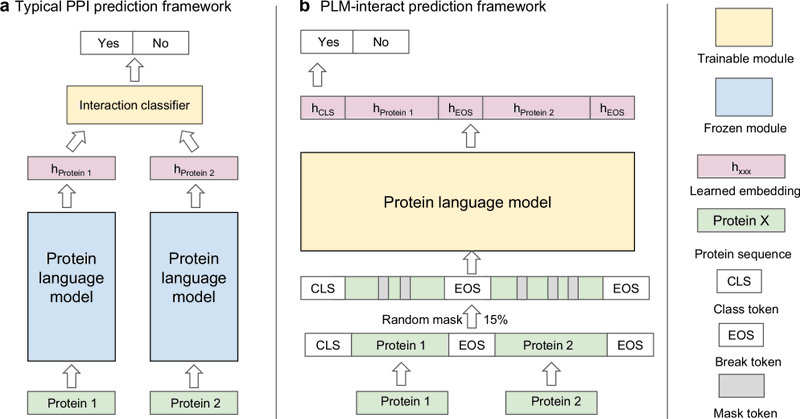
Our PLM-interact is out in @NatureComms! We show that jointly encoding protein pairs using protein language models improves protein–protein interaction prediction performance and enables fine-tuning to predict mutation effects in human PPIs. https://t.co/9unrZqULUS
nature.com
Nature Communications - Protein structure can be predicted from amino acid sequences with unprecedented accuracy, yet the prediction of protein–protein interactions remains a challenge. Here,...
1
3
16
Scientists @UofGlasgow have harnessed a powerful supercomputer, normally used to study the universe, to develop a new machine learning model which can help translate the language of proteins. Read more about the new @NatureComms study ➡️ https://t.co/kLWGk9J3Bg
1
13
21
Preparing for an evolving potential pandemic virus requires novel approaches to vaccine design. Fabulous work by Mathilde Richard’s team using central antigen design showing broad protection from antigenically diverse H5 avian influenzaviruses https://t.co/uJ9Ycf8RrY
@ErasmusMC
nature.com
Nature - A high-resolution antigenic map of influenza A(H5) haemagglutinin (HA) enables the design of immunogenic and antigenically central vaccine HA antigens that elicit antibody responses...
5
15
43
Earlier this year, we welcomed @DoctorChrisVT & the @STVStudios team to the CVR to chat about our ground-breaking work in #pandemic preparedness, virus discovery and using AI for protein prediction. Watch @BBC's Disease X: Hunting the Next Pandemic here: https://t.co/svbVVw7Rbq
0
4
12
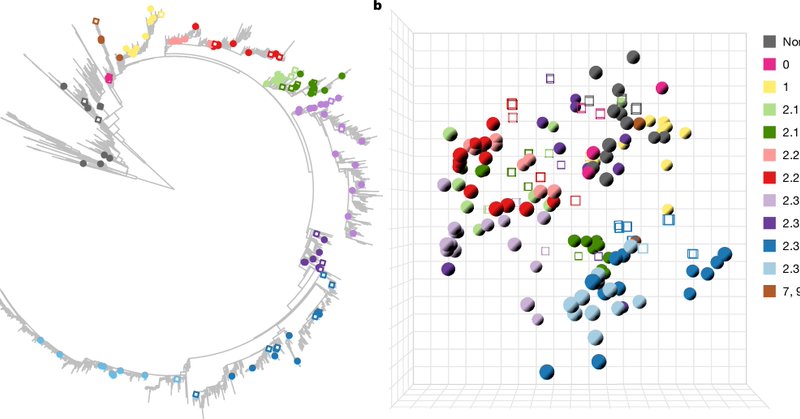
📢 NEW | The potential of H5N1 viruses to adapt to bovine cells isn’t uniform—it varies throughout their evolution. An international study led by CVR reveals how the potential to cross the species barrier varies during the evolution of H5N1 viruses. https://t.co/lnKedvS8fu
0
3
8
A warm welcome to our new Principal and Vice-Chancellor, Professor Andy Schofield. Prof Schofield shares a message for the UofG community. Here’s to this next chapter 💙 #TeamUofG #UofG #Glasgow #University #GlasgowUni #UniversityofGlasgow #GlasgowUniversity #scotland
18
29
119
This year’s medicine laureate Shimon Sakaguchi discovered a new class of T cells. Sakaguchi was swimming against the tide in 1995, when he made a key discovery. At the time, many researchers were convinced that immune tolerance only developed due to potentially harmful immune
34
1K
4K
We are at our future planning meeting at main campus of University 🏰 @CVR_Genomics @CVRbioinfo
0
3
16
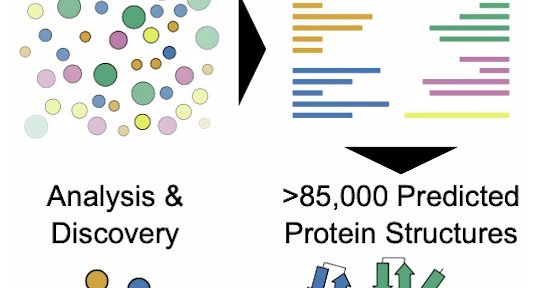
New exciting resource published!! The Viro3D paper is out, describing our comprehensive database of predicted virus protein structures! 💻🧬 Work with @CVRinfo 's Ulad Litvin, Joe Grove, Alex Jack, @robertson_lab and @blJOg
https://t.co/oQ0y5pDLcd
link.springer.com
Molecular Systems Biology - Viruses are genetic parasites of cellular life. Tolerance to genetic change, high mutation rates, adaptations to hosts, and immune escape have driven extensive sequence...
2
17
39
Our new study is out at @CellReports! We used 53 sarbecovirus pseudoviruses and 17 ACE2 expressing cells and demonstrated that 4 residues in spike contribute to the expansion of ACE2 usage range. Congrats Yusuke🎊! https://t.co/dLH5uZmmHD
cell.com
Kosugi et al. elucidate sarbecovirus evolution and ACE2 receptor tropism in natural hosts (12 Rhinolophus bat species), potential intermediate hosts (civets and raccoon dogs), and humans using a...
1
15
54