Dan Liu
@DanLiu_
Followers
137
Following
98
Media
9
Statuses
36
Computational biologist | bioinformatics, protein language models, virus-host interactions, LLMs 🦠 💻
Glasgow, Scotland
Joined May 2017
Our PLM-interact is out in @NatureComms! We show that jointly encoding protein pairs using protein language models improves protein–protein interaction prediction performance and enables fine-tuning to predict mutation effects in human PPIs. https://t.co/9unrZqULUS
nature.com
Nature Communications - Protein structure can be predicted from amino acid sequences with unprecedented accuracy, yet the prediction of protein–protein interactions remains a challenge. Here,...
1
3
16
📢 NEW | Introducing PLM-interact: a new AI-powered protein language model to predict protein-protein interactions Read the article: https://t.co/mOPm5IgN6A Find out more in the mini-podcast:
0
2
4
PLM-interact is out! We learned a lot along the way, from ColBERT to next sentence prediction for PPI, from zero short PPI mutation effect prediction to full model fine-tuning, from not knowing FSDP to burning 30k GPU hours in just a few days. Heroic effort from @DanLiu_
Our PLM-interact is out in @NatureComms! We show that jointly encoding protein pairs using protein language models improves protein–protein interaction prediction performance and enables fine-tuning to predict mutation effects in human PPIs. https://t.co/9unrZqULUS
0
2
11
A huge thanks to @craig_macdonald, @robertson_lab, and @keyuan1 for supervising this work, and to all my co-authors @FranYoung5, @Kieran12Lamb, @Adalberto_Cq, Alexandrina Pancheva, and Crispin Miller.
1
0
6
To summarise, PLM-interact extends single-protein PLMs to jointly encode interacting partners. It achieves state-of-the-art performance in cross-species and virus–host PPI prediction tasks and can be fine-tuned to predict mutation effects in human PPIs.
1
1
1
PLM-interact was applied to virus–human PPI prediction. The model outperforms existing approaches, achieving 5.7%, 10.9%, and 11.9% gains in AUPR, F1, and MCC, respectively — effectively capturing virus–host interactions at the protein level.
1
1
1
We further demonstrate examples where PLM-interact correctly predicts the effects of mutations on PPIs associated with human diseases. These results highlight its potential to identify whether disease-associated mutations weaken or strengthen protein interactions.
1
1
1
We fine-tuned PLM-interact to predict the effects of mutations on protein interactions — identifying whether mutations increase or decrease interaction strength. The fine-tuned model significantly outperforms zero-shot PPI models in the mutation-effect prediction task.
1
1
1
PLM-interact achieves state-of-the-art performance on a widely adopted cross-species PPI prediction benchmark — trained on human data and tested on mouse, fly, worm, yeast, and E. coli.
1
1
1
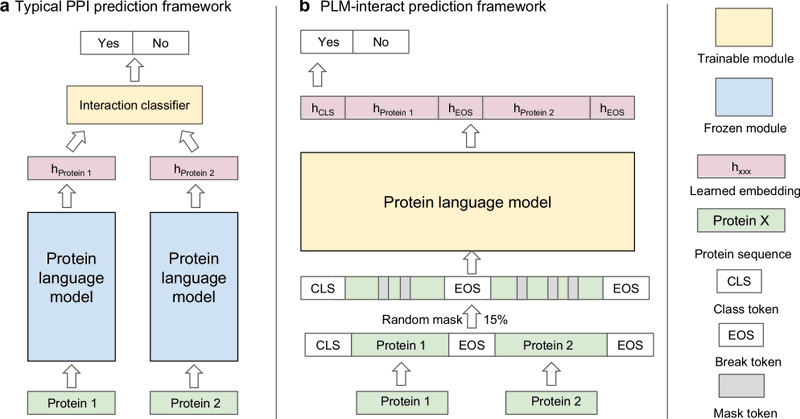
Existing PPI models use pre-trained PLMs to embed each protein separately, ignoring amino acid interactions between proteins. PLM-interact goes beyond single-protein encoding by jointly representing protein pairs to learn their relationships.
1
1
1
Protein language models trained on massive protein sequence datasets capture evolutionary, sequence and structural features — becoming the method of choice for representing proteins in state-of-the-art PPI predictors.
1
1
1
This work is part of my @viroinf PhD research, carried out under the supervision of @craig_macdonald, @robertson_lab, and @keyuan1, with HPC support from @DiRAC_HPC.
1
0
2
EvoMIL uses protein language models & deep learning to predict virus-host associations with improved accuracy & highlights critical viral proteins. #VirusHostInteractions #DeepLearning #Bioinformatics 📄 https://t.co/0IpmAXAhWx EVBC👤: @robertson_lab
journals.plos.org
Author summary Being able to predict which viruses can infect which host species, and identifying the specific proteins that are involved in these interactions, are fundamental tasks in virology....
0
3
10
Prediction of virus-host associations using protein language models and multiple instance learning @PLOSCompBiol 1. EvoMIL introduces an innovative method for predicting virus-host associations by combining protein language models (PLMs) and attention-based multiple instance
0
5
17
Big news: We just released PLM-interact, a tool for predicting protein-protein interactions, showing a 16-28% improvement over previous methods and even predicting mutation effects on interactions. Here’s the story behind this journey. 🧵👇
🚀 Our new preprint is out! We show that protein language models can predict protein-protein interactions by jointly encoding protein pairs, leading to significant improvements in PPI prediction. https://t.co/i81iRVZAls
1
12
26
Can protein language models be adapted to accurately predict protein-protein interactions across different species and mutation scenarios? @UofGlasgow @biorxivpreprint "PLM-interact: extending protein language models to predict protein-protein interactions" • The prediction
0
4
9
PLM-interact: extending protein language models to predict protein-protein interactions 1. PLM-interact introduces a novel approach to predict protein-protein interactions (PPIs) by jointly encoding protein pairs, leveraging a method similar to “next sentence prediction” in NLP.
1
5
27
A huge thanks to @FranYoung5, @Kieran12Lamb, @Adalberto_Cq, Alexandrina Pancheva, Crispin Miller, @craig_macdonald, and my supervisors @keyuan1, @robertson_lab.
0
0
6