The Bustamante Lab
@BustamanteLab
Followers
766
Following
61
Media
1
Statuses
28
Prof. Carlos J. Bustamante's Single-molecule Biophysics Laboratory at UC Berkeley
Berkeley, CA
Joined July 2019
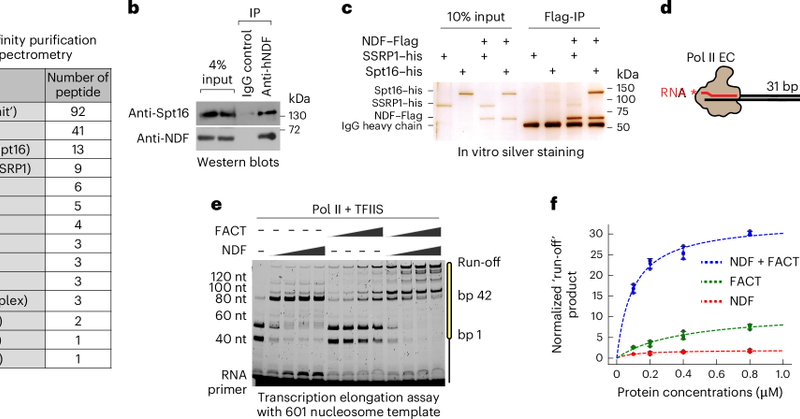
Excited to share our new @NatureCellBio paper with the Fei lab! Congrats to @Franburgos_, Chen Li, and Alex Tong. We show NDF–FACT condensates help Pol II traverse nucleosomal barriers. https://t.co/0EtGGWYlID
#TranscriptionChromatin #LLPS #SingleMolecule @LUMICKS_nl
nature.com
Nature Cell Biology - Li, Burgos-Bravo and colleagues report that NDF phase separation regulates FACT condensation, which enhances transcription by generating a localized biochemical environment...
0
2
16
Targeting FACT in cancer cells has shown efficacy in preclinical trials, but by which mechanism? Unclear. Enter: this new study from the @BustamanteLab at UC Berkeley. It reveals that NDF-FACT condensates form on transcribing PolII and enhance nucleosome disassembly and PolII
0
3
6
So proud of the smashing success of the International Course in Biophysics co-organized with my @berkeleyMCB colleague Carlos Bustamante @BustamanteLab and University of Salamanca @usal
https://t.co/VlCe4Pl20M Course ends with a Keynote from Nobel Laureate Richard Henderson🥇
biophysal.org
First Summer International Advanced Biophysics and Structural Biology Course. Eva Nogales - Carlos Bustamante.
0
10
36
Excited to share our new paper in @MolecularCell! Congratulation to the transcription team @Franburgos_ AlexTong, ChenLi, @CesarDiazC and our fantastic collaborators @ReinbergDanny & Kaplan labs! @BustamanteLab
#Transcription #Chromatin #SingleMolecule #OpticalTweezers #FACT
Online Now: FACT weakens the nucleosomal barrier to transcription and preserves its integrity by forming a hexasome-like intermediate
1
5
28
Check out our recent article in collaboration with the Fan lab: @BustamanteLab work led by @Honglu_Zhang in @ScienceAdvances! Congrats to all authors involved! @cccDNA
https://t.co/tI8X9dUhTD
science.org
DNA origami force probes and AlphaFold combine experiments and AI to decode the dynamics of protein folding.
0
4
13
Our last nucleosome work in collaboration with the Ren group: Nucleosome arrays visualized by individual particle cryo-ET. Congratulations @Meng3054 , @CesarDiazC , Jianfang Liu, Jinhui Tao, Paul Ashby, Carlos , and Gary Ren!! https://t.co/jIEG2q7d8r
nature.com
Nature Communications - Here, using cryo-ET, the 3D structures of individual nucleosome particles were characterized to observe changes under varying ionic strengths and in the presence of protein...
0
5
18
Our last work on nucleosome disassembly using High-speed AFM. Can we visualize the nucleosome plasticity using AFM imaging? Congratulations to Bibiana Onoa and the nucleosome team!! @cccDNA @CesarDiazC and Antony Lee. https://t.co/pmt2lsXqmM
Witnessing the unraveling of chromatosomes and nucleosomes in real-time with AFM! The intricate stepwise disassembly reveals a mechanism for unlocking DNA for vital cellular processes. #DNAUnlock#AFM NEW #ASAP Read it here: https://t.co/2rOKLKNowf
0
6
54
I am beyond excited that our story where we address the modalities of membrane tension propagation in cells is out @CellCellPress ! https://t.co/DJFSX5KYuX This is the fruit of an amazing collaboration with @OWeinerLab @BustamanteLab @virtual_embryo
12
50
206
Very!!! exciting news from the labs of Profs. Ting Xu, Carlos Bustamante and Haiyan Huang @TingXu_Berkeley @BustamanteLab
news.berkeley.edu
Using AI, Ting Xu and her colleagues designed polymer mixtures that mimic the natural proteins in biological fluids. The technique could improve the design of biocompatible materials.
0
2
9
Check out this this wonderful collaboration with @BustamanteLab . Congratulations to our biophysics PhD student @AlfredoFlorez07 and the rest of the team!
cell.com
A combination of bulk, single-molecule, high-throughput sequencing, and cryo-EM demonstrates that the ribosome, through the action of mechanical force and allostery, enhances the activity and reduces...
1
27
85
This semester's Steenbock Lectures will be given by Carlos Bustamante @UCBerkeley. Bustamante's lab focuses on single molecule #biophysics. More info: https://t.co/n74DMzMPxi.
@cijilim @UWMadisonCALS @UWMadBiophysics
0
3
16
Really hyped to share this preprint of some of my postdoctoral work with @OWeinerLab @virtual_embryo @BustamanteLab in which we set out to investigate whether membrane tension propagation in cells https://t.co/6L8ZzusTky Here is a summary of our findings 🧵
biorxiv.org
Membrane tension is thought to be a long-range integrator of cell physiology. This role necessitates effective tension transmission across the cell. However, the field remains strongly divided as to...
12
44
155
New Review! “The Development of Single Molecule Force Spectroscopy: From Polymers Biophysics to Molecular Machines” #Biophysics #SingleMolecule
The Development of #Single_Molecule_Force_Spectroscopy: From Polymer Biophysics to #Molecular_Machines
@BustamanteLab
https://t.co/216uhiVYI1
0
2
29
Hola, our latest work is now up on PNAS. Mechanical disassembly of nucleosomes using optical tweezers with fluorescence detection (Fleezers). @CesarDiazC, @cccDNA, Robert, Juan Pablo, @Meng3054, Enze, @AndyQuaen, Michael, Ryan, Bibiana, and Carlos. https://t.co/k85quIUkli
pnas.org
Nucleosome DNA unwrapping and its disassembly into hexasomes and tetrasomes is necessary for genomic access and plays an important role in transcri...
1
20
83
Hola, the final version of our latest paper is now up on Mol Cell. The question: how does nucleosome arrays form liquid droplets? By a two-step process. @Meng3054, @CesarDiazC, Bibiana, @cccDNA, @katy_requejo, Jianfang, Michael, Eva, Gary and Carlos. https://t.co/xjo1so7oAF
cell.com
Zhang et al. demonstrate that liquid-liquid phase separation of tetranucleosome arrays occurs in two steps: a spinodal decomposition involving formation of loosely arranged irregular condensates,...
1
27
90
Congratulations @kingcada!!
Ever stop and wonder how proteins cut membranes? It turns out this is actually a complicated question. There are many proteins that employ different mechanisms but for today, I'd like to focus on a set of proteins called ESCRTs. ( https://t.co/iha6Ggf9bM)
0
2
8
Hola everybody, we are excited to share our last preprint about nucleosome disassembly under force using optical tweezers combined with fluorescence detection. @CesarDiazC , @cccDNA , Robert, Juan Pablo.. #nucleosome #single-molecule #opticaltweezers
https://t.co/NBof4My1wr
biorxiv.org
Nucleosome DNA unwrapping and its disassembly into hexasomes and tetrasomes is necessary for genomic access and plays an important role in transcription regulation. Previous single-molecule mechani...
1
19
48
El Viernes 3 de Diciembre Juan P. Castillo, @BustamanteLab, va a presentar en el Journal Club de la SoBLA su artículo en @NatureComms "A DNA packaging motor inchworms along one strand allowing it to adapt to alternative double-helical structures" Regístrate: sobla.biof@gmail.co🧬
2
8
9
Dear community, we are excited to share our preprint about the 3D molecular architecture of nucleosome liquid droplets. Congrats to the authors of this work: @Meng3054, @CesarDiazC, Bibiana, @cccDNA, @katy_requejo, Michael, Eva, Gary and Carlos. https://t.co/rjKXFxvx5P
biorxiv.org
It has been proposed that the intrinsic property of nucleosome arrays to undergo liquid-liquid phase separation (LLPS) in vitro is responsible for chromatin domain organization in vivo . However,...
0
11
51
@zhijie_berkeley and Alan's collab w/ Quan Wang's lab is published in PNAS! Congrats to all authors! "Single-molecule diffusometry reveals no catalysis-induced diffusion enhancement of alkaline phosphatase as proposed by FCS experiments" @BustamanteLab
pnas.org
Theoretical and experimental observations that catalysis enhances the diffusion of enzymes have generated exciting implications about nanoscale ene...
0
4
10