Michael J. Robertson
@BiophysicsGuy
Followers
558
Following
1K
Media
26
Statuses
261
Assistant Prof of Biochem & Mol Pharm, Baylor College of Medicine. CryoEM, Computational Biophysics, Drug Discovery. Stanford/Yale/Michigan Alum.
Joined November 2014
Excited to share our latest preprint! We used cryoEM and MD simulations to look at the GTP-induced activation of the mu-opioid receptor-Gi complex with partial, full, & super agonists resolving Gi activation and how its modulated by different ligands! https://t.co/2E5cQPf6Xb
1
21
131
Happy to share that our paper on Metric Ion Classification (MIC) is now published! https://t.co/Wt852DcPVz
1
0
7
The Robertson lab has multiple openings for postdocs! Particularly interested in those with experience in either plate reader assays or ML, but all are welcome!; come learn about GPCRs, cryoEM, MD/FEP, drug discovery and more! https://t.co/LXa6IYHM7O
0
4
20
We are recruiting a young researcher to join our dynamic lab at UCSF - this is a great opportunity if you would like to get some research experience before graduate or medical school! https://t.co/jXVE2BoWlP. Please RT!
1
14
33
Woohoo, our paper on the mechanism of a G protein inhibitor that is specific to one family of G proteins is out! Nice collab with the Blumer nad @AndreaSoranno labs https://t.co/oUMFV8ZTcF
2
13
46
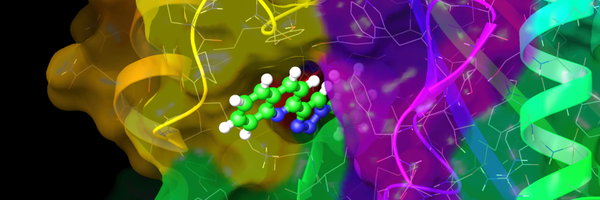
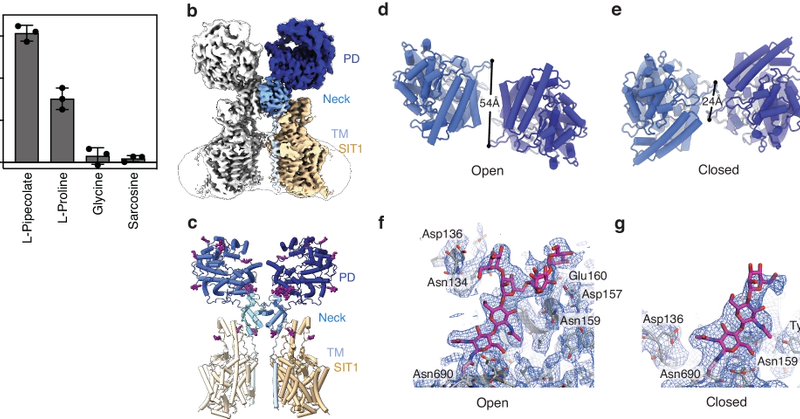
And it's out! Structure and function of the proline transporter SIT1 in complex with ACE2 Congrats to Huanyu, Ashley, Irina, @robertvandenbe2, Liz Carpenter, @T_dafforn, and the many other collaborators from @CmdOxford, @uobbiosciences, @Sydney_Uni
https://t.co/JkxTdmCxP6
nature.com
Nature Communications - The SIT1-ACE2 complex transports the amino acid proline and is the receptor of SARS-CoV-2. Here, the authors identify specific sequence requirements for proline transport...
1
11
54
Calling the #moleculardynamics community – first CASP target ever for predicting water (ensembles) around a macromolecule: https://t.co/HrYSuBFTMq Will be evaluated by direct comparison to #cryoEM on the Tetrahymena ribozyme #RNA!
1
11
39
"Mechanistic insights into G protein coupling with an agonist bound GPCR" Cool work led by the Hildebrand group.
linkedin.com
Hossein Batebi, Peter Hildebrand so happy 🎉 to see our work finally out in the open! What a long, wonderful ride (with ups and downs 🎢 ) with beautiful results in the end. It’s been great working...
1
1
8
1/6 Happy to share our new preprint- https://t.co/G1fDEnwCIi
@lab_stroud, @GuptaLab_, Jeffrey Bennett, @Nikk_click, Andrew Nelson, Peter Hwang, Sergei Pourmal. Huge thanks to the funding agencies @NIHgrants @guthyjacksonfdn @UCSF ( https://t.co/ByocobDoyS) for their support.
biorxiv.org
Neuromyelitis Optica (NMO) is an autoimmune disease of the central nervous system where pathogenic autoantibodies target the human astrocyte water channel aquaporin-4 causing neurological impairment....
2
7
28
Our paper is finally out in it's final form!
Online Now: The full spectrum of SLC22 OCT1 mutations illuminates the bridge between drug transporter biophysics and pharmacogenomics https://t.co/M8SDKeMgCJ
8
30
138
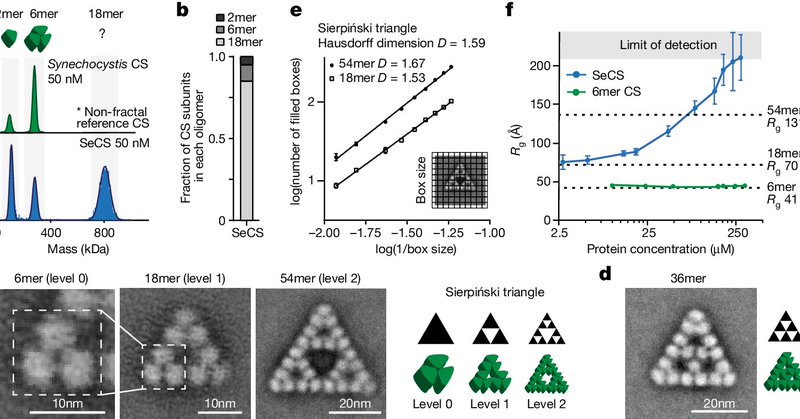
Alright everybody, what will undoubtedly forever be the lab's strangest discovery is now out in Nature. It's a protein that evolved to self-assemble into a Sierpinski fractal. https://t.co/zV5CDzbaGm 1/n
nature.com
Nature - Citrate synthase from the cyanobacterium Synechococcus elongatus is shown to self-assemble into Sierpiński triangles, a finding that opens up the possibility that other naturally...
15
197
883
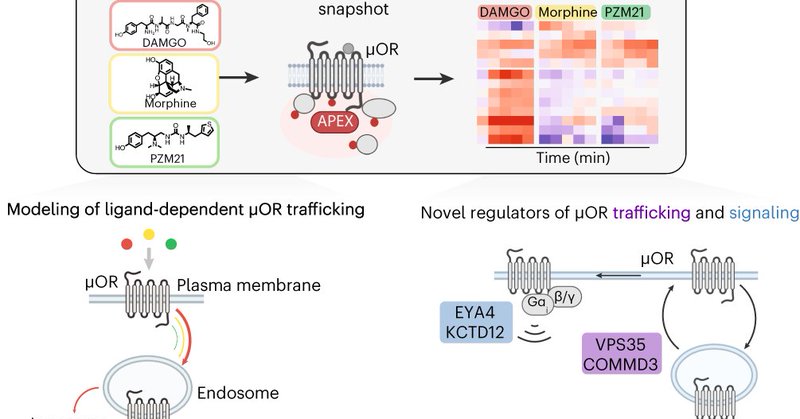
So excited to see our work on "Profiling the proximal proteome of the activated μ-opioid receptor" using APEX-based proximity labeling and quantitative mass spec published today. It is the result of an amazing team effort! https://t.co/e57PhS2DXe
#TeamMassSpec #Proteomics #GPCR
nature.com
Nature Chemical Biology - A proteomics and computational approach was developed to map the proximal proteome of the activated μ-opioid receptor and to extract subcellular location, trafficking...
8
40
173
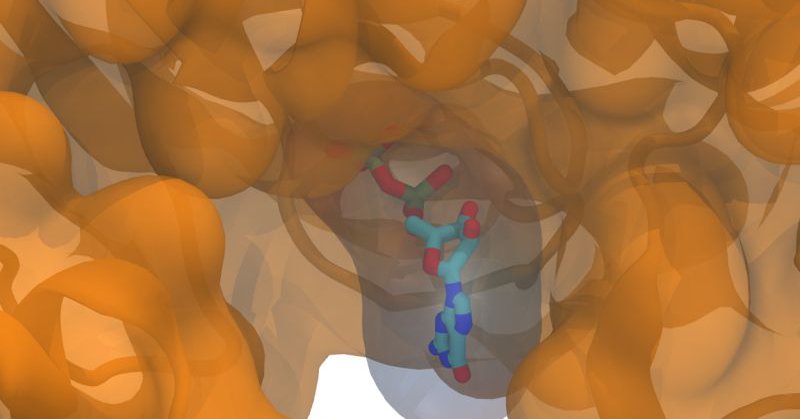
Very excited to share our new preprint where we develop MIC, a deep metric learning tool for classifying waters & ions in experimental structures; fantastic collaboration with the extremely talented Laura Shub and Selina Liu from the Keiser lab! https://t.co/hEl2snotyn
2
21
73
Our lab is planning an outreach activity for kids around proteomics. Besides the activity, we would love to show a short video clip how proteomics works eg how the proteins/peptides are measured in the mass spectrometer. Does anybody have a suggestion for a video? #TeamMassSpec
5
9
33
(1/7) Although I had the pleasure of bringing this manuscript to its final form released today, this work is built upon years of research by many people who deserve to have their contributions highlighted (a thread)...
Very happy to have my recent work on "Time-resolved cryo-EM of G protein activation by a GPCR" out in Nature today! #cryoEM #GPCR @Nature #LabSkiniotis
1
2
16
Very happy to have my recent work on "Time-resolved cryo-EM of G protein activation by a GPCR" out in Nature today! #cryoEM #GPCR @Nature #LabSkiniotis
10
65
271
Excited to announce a new preprint: https://t.co/eL6ReY20KK We developed a high-throughput method to measure the thermostability of RNA tertiary contacts using DMS chemical mapping and Mg2+ titrations. We believe this method is highly generalizable to many different systems. 1/
biorxiv.org
Structured RNAs often contain long-range tertiary contacts that are critical to their function. Despite the importance of tertiary contacts, methods to measure their thermodynamics are low throughput...
3
23
74
Excited to have my maps and models released this week from our time-resolved #cryoEM manuscript. Associated EMPIAR micrograph data sets coming soon, too! 😁
This week, we released a groundbreaking 160 entries from time-resolved #CryoEM capturing G protein activation by a #GPCR. It’s the largest data set from a single publication and describes the order of events driving G protein activation upon GTP binding!
1
5
36
We’re hiring postdocs and a lab tech! Experimental & computational people welcome; come learn about GPCRs, cryoEM, MD/FEP, drug discovery, method development and more! Feel free to DM me for more info. https://t.co/Um99mQKvZ5
https://t.co/RywVLPE24H Please RT!
1
31
51
Really fantastic work!
Excited to share our work published in @NatureComms. This was a joint collaboration between the Bernhard lab, @MoellerGroup and my lab with great help from the Kolb lab, @SteffenPockes, @HSchihada, @MPersechino_ and others. https://t.co/wpdSMgrdq4
0
0
4