Das Lab
@RDasLab
Followers
2K
Following
112
Media
37
Statuses
227
Our lab seeks an agile and predictive understanding of how RNAs structurally code for information processing and replication in living systems.
Stanford, CA
Joined April 2019
Nice overview in @naturemethods of #RNA structure approaches featuring our very own @RachaelKretsch and lots of amazing colleagues across the world
nature.com
Nature Methods - A steadily growing toolbox of techniques is helping researchers to understand the structural complexity that enables RNA molecules to perform a dizzying array of functions in the...
1
6
26
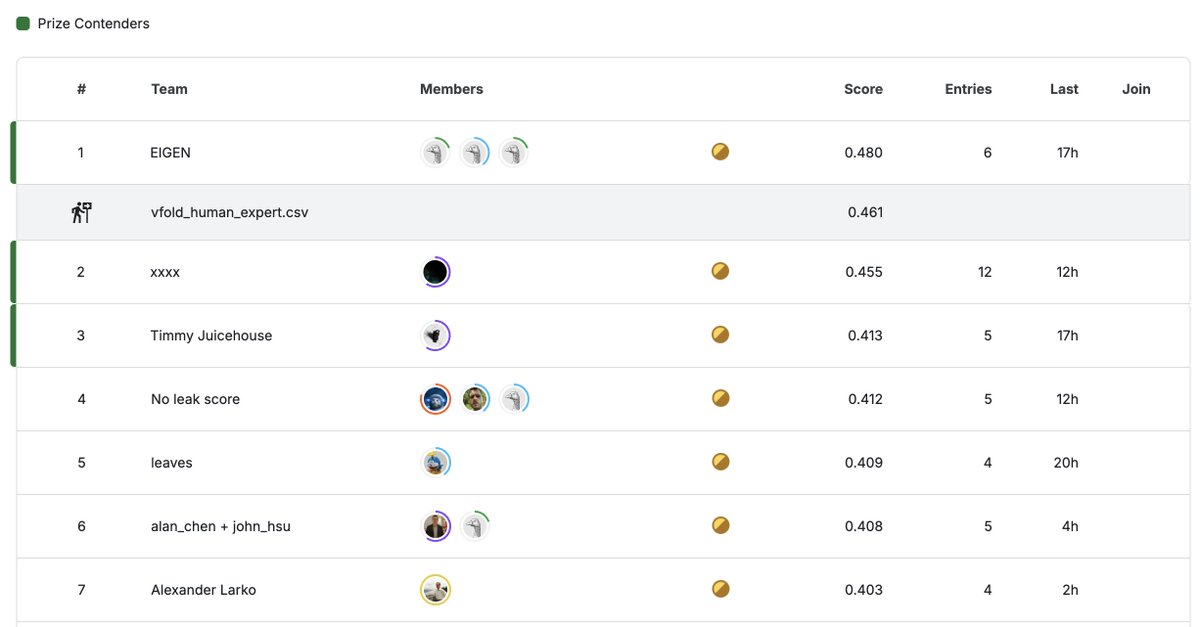
At Final Submission Deadline, numerous @kaggle notebooks outperform VFold human expert baseline in #RNA 3D structure prediction! Will these codes sustain performance on new targets released between now and Sep 2025? Join the discussion: https://t.co/6wMz2E1UK4
0
9
30
#RNA 3D structure prediction remains an open problem! Come test new ideas against SOTA & >1000 teams in Kaggle RNA 3D Folding competition, co-hosted by #CASP16 and @rnapuzzles, 2.5 weeks left. https://t.co/ODfoePDZ93 N/N
kaggle.com
Solve RNA structure prediction, one of biology's remaining grand challenges
0
0
2
#CASP16 enabled by 46 predictor labs, 22 experimental #RNA and #DNA groups, co-assessors @rachaelkretsch, @alissahummer, @ShujunHe0717, @RongqingY, Jing Zhang, Thomas Karagianes, @qiancong, @AKryshtafovych Many thanks! 7/N
0
0
0
Upon accounting for available templates, no clear improvement in #CASP16 #RNA compared to prior blind challenges #CASP15 @rnapuzzles, except automated servers are less poor than they used to be. 6/N
0
0
0
#AlphaFold 3 server outperformed by 8 groups — including another #CASP16 server. Human expert group Vfold is SOTA. 5/N
0
1
0
Nucleic acid assessment for #CASP16 blind competition is out. As in prior challenges #deeplearning for 3D #RNA structure in #CASP16 prediction remains disappointing. https://t.co/V5AZwux2zx 1/N
biorxiv.org
Consistently accurate 3D nucleic acid structure prediction would facilitate studies of the diverse RNA and DNA molecules underlying life. In CASP16, blind predictions for 42 targets canvassing a full...
7
15
52
Pretty new discovery! 💎 SLAC and @Stanford researchers using cryogenic electron microscopy showed for the first time that large RNA complexes can assemble without the help of proteins, expanding our understanding of RNA folding and function: https://t.co/f8Z59zDOc7
0
5
18
What are these ornate RNA structures doing in phages and bacteria? Can we use them for biotechnology? Great collab led by @rachaelkretsch, with Vivian Wu, Svetlana Shabalina, Hyunbin Lee, Grace Nye, Eugene Koonin, @algao8 and @chiulab 2/2
0
1
4
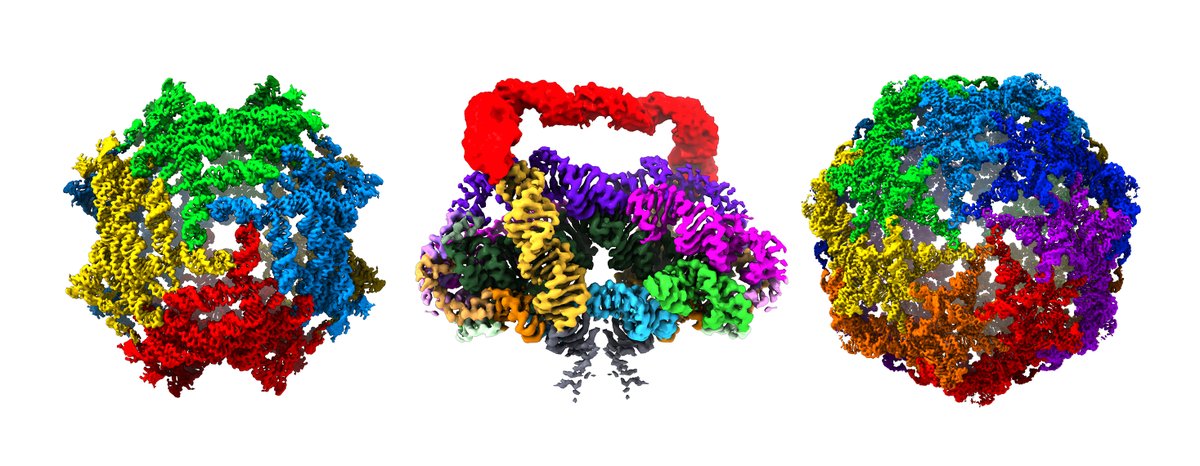
Our #RNA multimer paper out now in @nature! #cryoEM of 2 distinct nanocages GOLLD and ROOL and membrane-associated OLE. Multimers at ultra-low concentrations, special covariance for cross-molecule kissing loops, and speculation on function https://t.co/ZihCKHIIcX. 1/2
2
45
183
Stanford #RNA 3D Folding: With data refresh done, the first two superhuman submissions that make public their documented, no-cheat notebooks will get the Early Sharing Prizes. Will EIGEN be the first? https://t.co/3SiTmewCQc
@kaggle @HHMIJanelia @HHMINEWS @StanfordMed #CASP16
0
14
20
And many thanks to @LenaSteckelberg @HartmannLab @esandersen @chiulab @HoebartnerLab @cpricejones @KoiralaLab @cdmackereth @Marcia_lincRlab Joe Piccirilli @PhoebeARice @SuZhaoming and labs for their insightful analyses. Reality check on #RNA structure prediction! 2/2
0
0
0
Are RNA 3D models accurate enough to be biologically useful? #CASP16 experimentalists say: not so much. https://t.co/rUlFyDaiUJ Thanks for leading this paper @rachaelkretsch @AKryshtafovych! 1/2
biorxiv.org
Accurate biomolecular structure prediction enables the prediction of mutational effects, the speculation of function based on predicted structural homology, the analysis of ligand binding modes,...
2
15
49
@shujun @nvidia @kaggle @eternagame @nvidia blog on training Rnet2 with @SJ_He via NAIRR pilot program and DGX cloud -- more coming from this effort, including RNA design AI results on @eternagame -- stay tuned and do join the competition on @kaggle by May 2025:
developer.nvidia.com
The Das Lab at Stanford is revolutionizing RNA folding research with a unique approach that leverages community involvement and accelerated computing. With the support of NVIDIA DGX Cloud through the…
0
0
5
We see a path to scale up to a 30B param foundation model Rnet3 that should solve RNA 3D structure prediction. But #deeplearning training costs now dwarf data generation costs. Anyone have 100M GPU-h they can spare to solve one of biology’s remaining grand challenges? (N/N)
0
2
10
Much of the experimental tech driving Rnet2 — including ultrafast/cheap sequencing @UltimaGenomics and “AI-ready” Gentitan synthesis @GenScript — wasn’t even available last year. (6/N)
0
2
4
Rnet2 training data are on synthetic designs from RFdiffusion @UWproteindesign, gRNade@chaitjo, genome scans, and a new design method Shujun @SJ_He. Sequences mapped thanks to key experimental innovations by Ann Kladwang and Hamish Blair @rdaslab. (5/N)
0
0
0