Dr. Anna Ramisch
@AnnaRamisch
Followers
272
Following
2K
Media
1
Statuses
366
CompBio Postdoc interested in HumanGenetics, NeuroEpigenomics & Addiction @unige_en
Geneva, Switzerland
Joined December 2014
RT @aja_mys: 🔥🧬Finally out!🧬🔥 Check out our latest study in Nature Genetics uncovering a surprising phenomenon of convergent co-regulated p….
nature.com
Nature Genetics - Genome-wide analysis and genetic manipulation at loci regulated by p53, E2F4 and RFX7 show that convergent promoters with similar epigenetic features can be co-regulated and...
0
2
0
RT @kerrin_small: Very excited to share that our manuscript on cis-eQTL analysis of adipose tissue gene expression in 2,344 samples is now….
0
3
0
RT @clintomics: Delineating the effective use of self-supervised learning in single-cell genomics @NatMachIntell @fabian_theis https:/….
nature.com
Nature Machine Intelligence - Self-supervised learning techniques are powerful assets for enabling deep insights into complex, unlabelled single-cell genomic data. Richter et al. here benchmark the...
0
18
0
RT @Katrinardottir: I'm very excited to share that I’ve started as a group leader @karolinskainst. I am looking for curious, enthusiastic a….
0
21
0
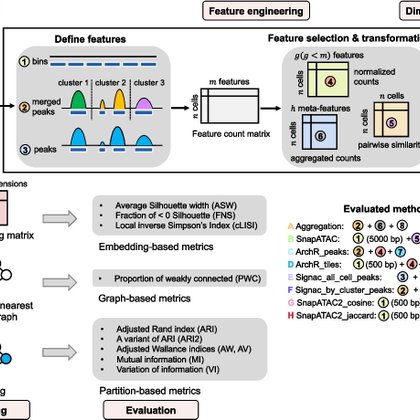
RT @clintomics: New benchmarking study of 8 pipelines and 5 widely used methods for scATAC data analysis. includes embedding, graph and par….
genomebiology.biomedcentral.com
Background Single-cell chromatin accessibility assays, such as scATAC-seq, are increasingly employed in individual and joint multi-omic profiling of single cells. As the accumulation of scATAC-seq...
0
36
0
RT @arnsbr: Really happy for @aline_real25 to be getting this out. Major implication for me is I've always assumed gwas hits will be explai….
0
1
0
So happy that our preprint is finally out! If you're interested in the advantages of long-read direct RNA sequencing in population studies, this is for you.
I am excited to share our new preprint with @anavinuela, @AnnaRamisch, @arnsbr, and @dermitzakis! We used direct RNA long-read seq on 60 LCLs from the 1000 Genomes Project to study genetic regulation of transcript abundance & RNA modifications (m6A).
2
2
17
RT @aline_real25: I am excited to share our new preprint with @anavinuela, @AnnaRamisch, @arnsbr, and @dermitzakis! We used direct RNA long….
0
14
0
RT @madisonahoner: ‼️New Epigenetics Review ‼️. Read here at @GenesDev where we discuss brief history of various epigenetic modulator class….
0
74
0
RT @smgbraun: Check out our lab's recent #preprint by Hanna Schwaemmle and our collaborators @unige_en. We asked how do cells assemble func….
biorxiv.org
The SWI/SNF (or BAF) complex is an essential chromatin remodeler that regulates DNA accessibility at developmental genes and enhancers. SWI/SNF subunits are among the most frequently mutated genes in...
0
27
0
RT @UNIGEnews: Des scientifiques de l’#UNIGE ont identifié les séquences génétiques régulant l’activité des gènes responsables de la croiss….
0
4
0
RT @EtienneSollier: Figeno, my visualization tool for genomics🎨🧬, is now published in Bioinformatics! .It can gener….
0
205
0
RT @guillaumeandrey: The revised version of our chondrogenic enhancer preprint is out today, featuring the in vivo assessment of enhancer d….
nature.com
Nature Communications - Chondrocyte differentiation controls skeleton development and stature. Here, the authors map mouse fetal chondrocyte enhancers, highlighting their role in controlling bone...
0
17
0
RT @evgenykvon: How do enhancers work over distances that sometimes exceed megabases? Excited to share our work led by @gracecbower where w….
0
232
0
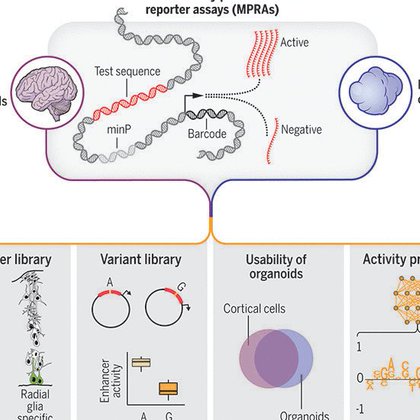
RT @NadavAhituv: Characterizing >100,000 sequences in primary neurons and organoids and deep learning to decode regulatory activity. Amazin….
science.org
Nucleotide changes in gene regulatory elements are important determinants of neuronal development and diseases. Using massively parallel reporter assays in primary human cells from mid-gestation...
0
21
0
RT @lcolladotor: Ana @anavinuela is motivating us to look at eQTLs beyond gene counts to transcript counts from long read sequencing data….
0
3
0
RT @tmontsay: Excited to share with the single-cell community the most recent work of @tmontsay and @AEsteveCodina from @cnag_eu, something….
biorxiv.org
The advent of droplet-based single-cell RNA-sequencing (scRNA-seq) has dramatically increased data throughput, enabling the release of a diverse array of tissue cell atlases to the public. However,...
0
23
0
RT @ZhenMiao2: Inspired by @lpachter and team's exploration of scRNA-seq pipelines, we face similar challenges in scATAC-seq, only magnifie….
0
79
0
RT @fabian_theis: Excited to share Nicheformer! Led by @Alejandro__TL & @AnnaCSchaar, Nicheformer is a foundation model for single-cell &….
0
90
0