Alexander Haglund
@AlexHaglund9
Followers
44
Following
39
Media
10
Statuses
33
Research Associate @ImperialBrains. Bioinformatics, causal inference and single-cell.
Joined November 2014
Very excited to share our latest publication in @NatureGenet! Our manuscript shows how Mendelian Randomization (MR) isolates putatively causal links between cell-type specific gene expression & brain phenotypes. https://t.co/ofuoFWphDs Tweetorial below 👇
2
5
16
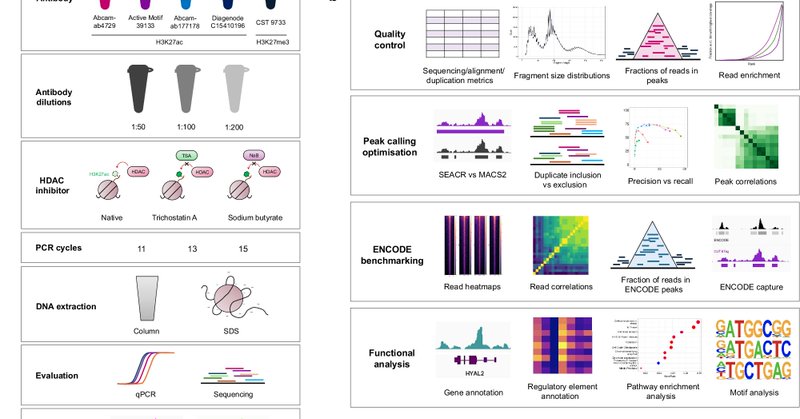
We're so thrilled to finally share with you the published version of our CUT&Tag optimization and benchmarking paper "CUT&Tag recovers up to half of ENCODE ChIP-seq histone acetylation peaks" - out in @NatureComms today:
nature.com
Nature Communications - Cleavage Under Targets & Tagmentation is a rapidly expanding technique, but thorough evaluation and benchmarking against established ChIP-seq datasets are required....
5
38
183
Just one day left to apply as computational genomics postdoc in our lab. Apply by tomorrow, 8 March: https://t.co/QU4f7H8Ci0
1
27
115
Exciting new role for a Computational Genomics Research Associate in the lab of @sj_marzi at the UK DRI at King's! Lead projects exploring the epigenome and transcriptome of brain cells in neurodegenerative diseases 🧠 🗓️Deadline: 08 March Full info👉 https://t.co/eQd1Jz8XiM
0
11
23
Plasma protein and brain structural imaging evidence that #SARSCoV2 is associated with greater brain β-amyloid pathology in older adults, particularly those hospitalized or with hypertension. https://t.co/CwY7tOYceR
@NatureMedicine a matched @uk_biobank case-control study
18
137
382
📢 Our lab is looking for a bioinformatics postdoc. We have lots of exciting projects to work on, including single cell data, microglia xenotransplantations and nanopore long reads splicing analysis. Look out for the official advert next week and get in touch in the meantime 🧬🧠
14
130
557
📢 CUT&Tag version 3 Sometimes the preprint to paper route can be quite the odyssey. And so in that vein, we are delighted to give you version 3 of our CUT&Tag optimization and benchmarking manuscript: https://t.co/OUgpMyG9F7
biorxiv.org
Techniques for genome-wide epigenetic profiling have been undergoing accelerated development toward recovery of high-quality data from bulk and single cell samples. DNA-protein interactions have...
2
39
179
Modern GWAS can identify 1000s of hits but it can be hard to turn this into biological insight. What key cellular functions link genetic variation to disease? I'm excited to present our new work combining associations + Perturb-seq to build interpretable causal graphs! A🧵
8
114
420
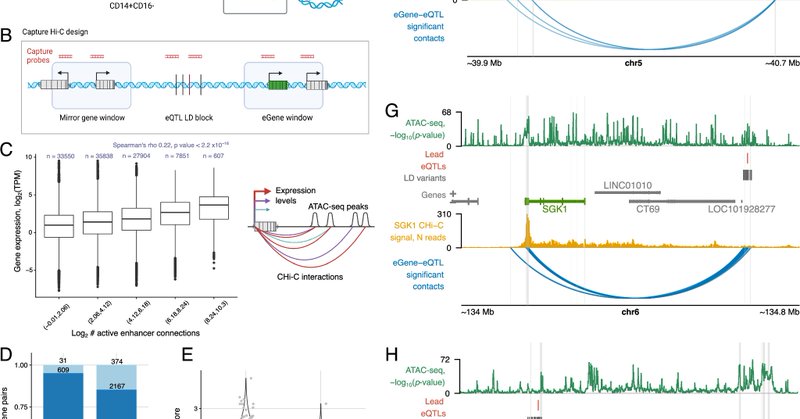
Our new paper is finally out, showing that the same enhancer sequences - and likely the same transcription factors recruited to them - affect the activity of distal enhancers and their looping over to target gene promoters. https://t.co/T1py66rC7h
nature.com
Nature Communications - Here, the authors study the effects of expression quantitative trait loci on enhancer activity and promoter contacts in primary monocytes isolated from male individuals,...
8
57
245
14/ What’s next? Landmark! The Landmark project is a public-private partnership involving GSK, Novartis, Roche and UCB with the hope to find novel targets for Parkinson’s Disease, currently the fastest growing neurological condition in the world. https://t.co/tVg3hYSyoS
imperial.ac.uk
Collaboration brings together charity, university and pharmaceutical sectors
0
1
3
13/ Big thank you to all co-authors including @VerenaZuber, @JulienBryois, @prashant1003, @AbouzeidMaya and many many more including important contributions from @DRI; @FancyNurun, @jojacksonhere and @nottalexi to name a few. Study conceived by Michael Johnson @ImperialBrains.
1
0
1
12/ Thank you for reading! Tons more not covered here in the publication! https://t.co/ofuoFWphDs. If you are interested to hear more, I will be presenting this work virtually at #ADPD2025 if you feel like dropping in!
6
0
3
11/ Why is this important? Replication at protein level confirms that inferences from gene expression are retained upon translation; in addition, pQTLs in other tissue (such as plasma) can provide very useful biomarkers for disease risk. Example with GPNMB COLOCs.
1
0
1
10/ Because these associations are expected to be mediated by a gene’s protein product, we repeated MR analysis using both UKB-PPP pQTLs and brain pQTLs (Robins et al.,). We replicated 11 in plasma and 20 in brain, including GPNMB, EGFR and CR1.
1
0
0
9/ For example, several genes were found to be putatively causal for PD risk; importantly, MR infers a directionality of effect (up or down) that can inform therapeutic targeting modelling. Example; increased expression of TMEM163 in microglia -> increased PD risk.
1
0
1
8/ Adhering to STROBE-MR guidelines, we selected robust independent eQTLs as instruments (F-statistic > 15) for MR, and found 140 putatively causal cell-type/gene/trait associations, many of which in Alzheimer’s Disease, Multiple Sclerosis and Parkinson’s Disease.
1
0
0
7/ MR necessitates associations free from disease presence to avoid reverse causation. Using a controls-only subset (N=183), we repeated eQTL + colocalization analysis. Despite a smaller cohort, we discovered an additional 91 COLOCs such as with PEX13 in MS (PP.H4 = 0.87).
1
0
1
6/ Many interaction-QTLs also influence colocalization; Notably, 23.6% of colocalizations showed disease dependency—e.g., TP53INP1 in the full cohort vs. controls-only.
2
1
2
5/ To uncover shared genetic regulation between gene expression and phenotypic risk, we employed genetic colocalization across 41 brain-related traits. We identified 501 COLOCs (PP.H4 > 0.8), with 74.4% specific to a single cell type.
1
0
1
4/ Combining disease and control tissue is common practice to maximize discovery power; however, we show that up to 41% of eQTLs interact with disease even after correcting for disease status. These relationships are important to clarify for downstream causal inference.
2
1
2
3/ An example below with GPNMB; this eQTL is specific to glia, where its strongest associations lie with astrocytes and OPCs but is absent from neurons, endothelial cells and pericytes. Why is this important? CT-specific eQTLs -> CT-specific causal inferences!
1
0
0