Yuan Qin
@yuanqinoffice
Followers
98
Following
135
Media
11
Statuses
51
🎓 Bioinformatics Ph.D. of CAS 📒 Editor of Journal of Genetics and Genomics (JGG) 👀 All views are my own
Beijing, China
Joined October 2022
RT @JGenetGenomics: Genomic characterization reveals distinct mutational landscape of acra. #East #Asian #WGS #A….
0
1
0
RT @EricTopol: The latest large language of life #AI model (LLLM) predicts the impact of coding and non-coding DNA variants with scores for….
0
157
0
SpaGRA: Graph augmentation facilitates domain identification for spatially resolved transcriptomics .🎉🎉🎉. 💻 >>> . 📖 >>> #SpatialTranscriptomics #DeepLearning #GraphNeuralNetworks.
github.com
Contribute to sunxue-yy/SpaGRA development by creating an account on GitHub.
0
0
0
KanCell: A deep learning model based on the Kolmogorov-Arnold network that integrates scRNA-seq and ST data to reveal cellular heterogeneity and spatial patterns💻. >>> #scRNAseq #SpatialTranscriptomics #DeepLearning
0
0
0
RT @EvomicsLab: The code of NanoTrans can be freely accesible via @github at
github.com
An integrated computational framework for comprehensive transcriptome analyses with Nanopore direct-RNA sequencing data - yjx1217/NanoTrans
0
1
0
#NanoTrans from @EvomicsLab, is an integrated computational framework for Nanopore direct RNA sequencing data analysis ⚡️⚡️⚡️. 📖 (Open Access): 🔧: #nanopore #DRS #longread #JGG.
github.com
An integrated computational framework for comprehensive transcriptome analyses with Nanopore direct-RNA sequencing data - yjx1217/NanoTrans
1
2
10
RT @EvomicsLab: Glad to see our NanoTrans paper out @JGenetGenomics.We developed an all-in-one platform for @nanopore direct RNA sequencing….
0
12
0
shinySRT, a new tool is online✨designed for the sharable and interactive visualization of spatially resolved transcriptomics data.#shinySRT #SRT #opensource #JGG.Get it & Detailed Tutorial: 📖:
github.com
Contribute to silhouette99/shinySRT development by creating an account on GitHub.
0
0
2
A new tool is online~.shinyTempSignal: an R shiny application for exploring temporal and other phylogenetic signals .#Molecularclock #shinyTempSignal #JGG #ggtree. Congrats and many thanks to @guangchuangyu . 🥳Get it: 📖Tutorial:
github.com
Explore Temporal and Other Phylogenetic Signals. Contribute to YuLab-SMU/shinyTempSignal development by creating an account on GitHub.
0
6
23
RT @JGenetGenomics: shinyTempSignal: an R shiny application for exploring temporal and oth. #Molecular clock #Ro….
0
2
0
RT @biorxiv_bioinfo: NanoTrans: an integrated computational framework for comprehensive transcriptome analyses with Nanopore direct-RNA seq….
0
6
0
RT @JGenetGenomics: JGG is now inviting submissions for a special issue of Zebrafish Biology! Submissions of original Research/Resource/Met….
0
5
0
Cover: two individuals in a debate over various metrics for evaluating the quality of genome assembly, struggling to reach a consensus. It underlines the significance of tools capable of comprehensive genome assembly evaluation. 🔧GAEP:
github.com
A genome assembly assessment pipeline. Contribute to zy-optimistic/GAEP development by creating an account on GitHub.
Issue 10 of JGG is online now. Cover story: Zhang et al. develop GAEP, a comprehensive genome assembly evaluating pipeline, which integrates several commonly used methods and tools for genome assembly evaluation, and introduce two newly developed modules.
0
0
5
RT @PanktiMehta24: 🧐The what, how, where & why of how antibodies or magic bullets are used in medicine ! 🪄 🔫. 🔹IV IG or intravenous immunog….
0
31
0
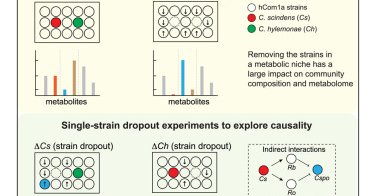
RT @mfgrp: Today we report that removing a strain from a complex microbiome can ripple through the community in an unpredictable (& useful)….
cell.com
Construction of variants of a complex defined microbial community reveals that strains that are functionally redundant within a niche can have widely varying impacts outside the niche.
0
103
0
RT @JGenetGenomics: GAEP: a comprehensive genome assembly evaluating pipeline #Genome assembly #Assembly evaluation….
0
4
0