Sara Bakić
@sarrabakic
Followers
69
Following
968
Media
4
Statuses
37
PhD student at @NUSingapore, @astar_gis | Bioinformatics | NLP | ML
Joined May 2022
I am happy to introduce Campolina, a deep neural framework that replaces traditional algorithmic approaches for nanopore signal segmentation and improves segmentation quality for real-time analysis. Preprint and details in the thread👇 K. Friganovic, @BryanHooi1, @msikic 1/7
2
8
15
Happy to present the first complete genome of an Indian individual https://t.co/2M92EvPBhw. Joint effort by @astar_gis ,FER, and partners, with support from the Singapore NPM programme! Joint effort with JJ Liu Lab! @msprasad693 and @JosipaLipovac as co-first authors.
biorxiv.org
Human reference genomes have been instrumental in advancing genomic and biomedical research, but South and Southeast Asian populations are underrepresented, despite accounting for a large proportion...
1
18
67
Campolina is fully open source with source code available at the GitHub repository https://t.co/fEOW1pcvid, and R9.4.1 and R10.4.1 weights available at Zenodo: https://t.co/p9nmShGTji. 7/7
zenodo.org
This repository contains trained models for Campolina, a deep learning framework for event border detection in nanopore sequencing signals. We provide models trained for R10.4.1 (R10_model.pth) and...
0
1
4
Furthermore, Campolina’s segmentation integrates seamlessly into existing real-time frameworks, improving signal classification results in multiple downstream tasks without any pipeline optimization while remaining computationally efficient. 6/7
1
0
3
and show that Campolina's segmentation quality is significantly higher than that of the traditional algorithm, Scrappie. The predicted borders match the ground truth ones better, and the obtained event levels correspond better to the expected levels on the reference. 5/7
1
0
3
To ensure a complete assessment of segmentation quality, we go beyond border matching by proposing a novel comprehensive evaluation pipeline that compares the predicted and ground-truth border positions, but also the obtained event levels and the expected reference levels 4/7
1
0
3
@BryanHooi1 @msikic Campolina is a convolutional neural network trained in a supervised manner to identify event borders in raw nanopore signals, with ground truth extracted through a refining signal-to-reference alignment framework. 3/7
1
0
3
@BryanHooi1 @msikic Campolina is the first deep learning model tailored for raw signal segmentation and the first segmentation framework developed for the R10.4.1 chemistry: https://t.co/N9gJ1vJsim 2/7
biorxiv.org
Nanopore sequencing enables real-time, long-read analysis by processing raw signals as they are produced. A key step, segmentation of signals into events, is typically handled algorithmically,...
1
1
3
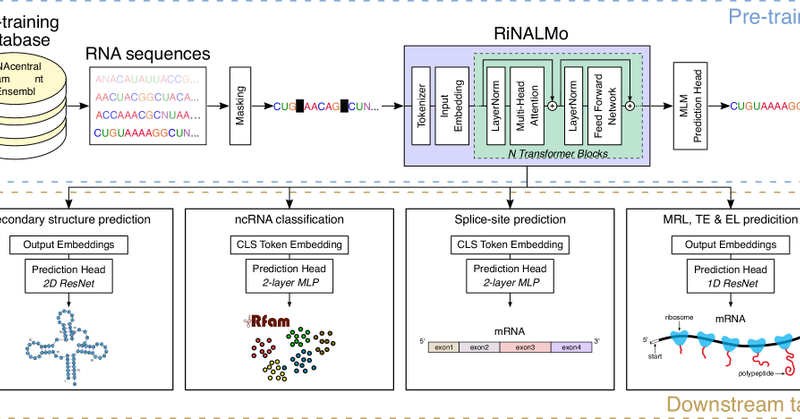
Our RiNALMo RNA language model has been published in Nat Comm https://t.co/2EXzExKdzK Great work by @RJPenic and @TinVlasic, with support and patience in teaching us RNA biology from Roland G Huber and @ywan_wan RiNALMo is already an SOTA as a benchmark for RNA LLMs.
nature.com
Nature Communications - RiNALMo, a large-scale RNA language model trained on non-coding RNA sequences, captures structural information and achieves state-of-the-art performance on multiple tasks,...
3
16
57
I am happy to share our new preprint introducing MADRe - a pipeline for Metagenomic Assembly-Driven Database Reduction, enabling accurate and computationally efficient strain-level metagenomic classification. @msikic, @r_vicedomini, @KrizanovicK 🔗 https://t.co/YUq9Xbp2Lm 1/9
biorxiv.org
Strain-level metagenomic classification is essential for understanding microbial diversity and functional potential, but remains challenging, particularly in the absence of prior knowledge about the...
4
13
12
GNNome was published in @genomeresearch! This is a novel paradigm for de novo genome assembly based on GNNs. Without explicitly implementing any simplification strategies, it can achieve results comparable or higher than other SOTA tools. Paper, code, and overview are 👇 [1/8]
4
41
132
🚨 We have extended the deadline for submissions! 🚨 📅 New deadline: Feb 13, 2025, AoE - so you can finish just in time for Valentine's day 🥰 🔗 More details on our website (link in bio)
0
7
10
We are organising https://t.co/bmP3bmhJHq at#ICLR2025. Topics: RNA, DNA and cell LLMs, structure, modifications, correction, variant calling... Deadline: Feb 10th 2025 Time for revision: 1 month Accepted papers - the opportunity to be invited by Nature Methods for submission
ai4na-workshop.github.io
AI4NA Workshop @ ICLR'25
0
8
19
🚨Exciting news! We're partnering with @naturemethods 🧬High-quality submissions to the AI4NA workshop on @iclr_conf can also be considered for publication in the journal! 🌟 👉More details on our web page (link in comments) #AI4NA #ICLR2025
2
5
7
📜 Did you know that, apart from the track for full papers, we also have a track for tiny papers that are up to 3 pages long? As part of @iclr_conf initiative, first-time submitters and authors from underrepresented minorities can opt for this track. Details in the comment 👇
1
6
10
📢 Submissions are open! Join us at #AI4NA 🇸🇬 @iclr_conf to present your latest work 🧬 🚨 Updates: We've revised paper length guidelines & extended the deadline to Feb 10, 2025, AoE. 🔗 Details on our webpage (link in comments). Don’t miss out! 🌟
1
4
6
🚨 1 month to go! 🚨 The submission deadline for the AI4NA workshop at @iclr_conf is fast approaching! 🧬 ✨ Submissions on OpenReview will open soon—stay tuned! ✨ 🔗 Learn more on our web page (link below 👇) #AI4NA #ICLR2025
1
6
9
🚀Thrilled to be part of ICLR 2025! Join our workshop AI for Nucleic Acids (AI4NA) to explore cutting-edge research and connect with field leaders. Thanks to our organizers and @iclr_conf for making this possible. Find the link to the website in our bio! More info below👇
1
6
10
Our hitchhiking paper is out at https://t.co/CX6347vrmF - a great collaborative effort with @msprasad693 @JosipaLipovac, @FilipTomas95 JJ Liu. Briefly, @nanopore is enough for high-quality human genomes. Recently, HERRO showed that even UL is sufficient. 1/3
link.springer.com
Genome Biology - Long-read technologies from Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT) have transformed genomics research by providing diverse data types like HiFi,...
2
21
41