Lovro Vrček
@lovrovrcek
Followers
378
Following
3K
Media
34
Statuses
235
PhD student @fer_unizg | Bioinformatics Specialist @astar_gis. AI, graphs, bioinformatics. 🇭🇷
Singapore 🇸🇬
Joined October 2018
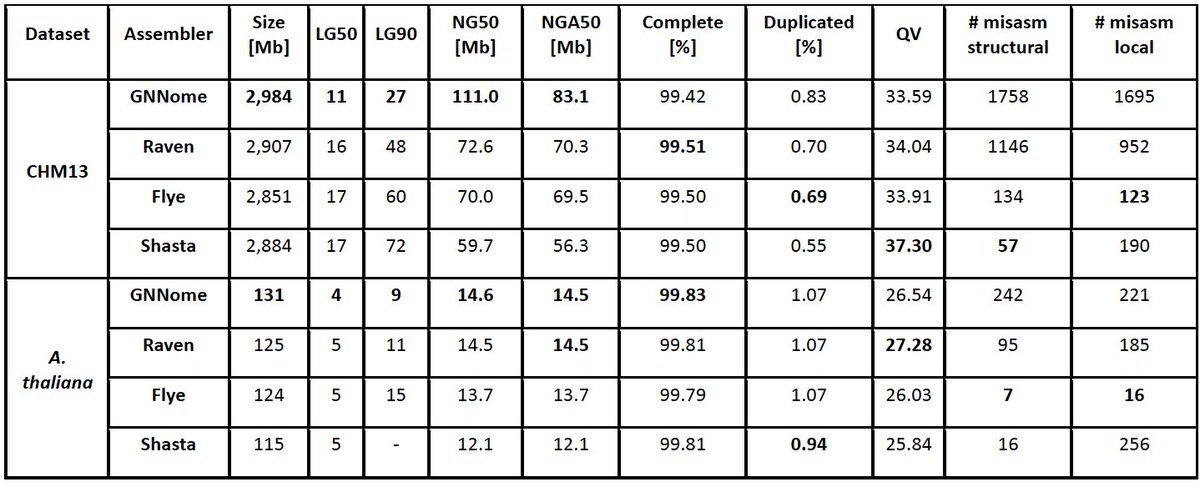
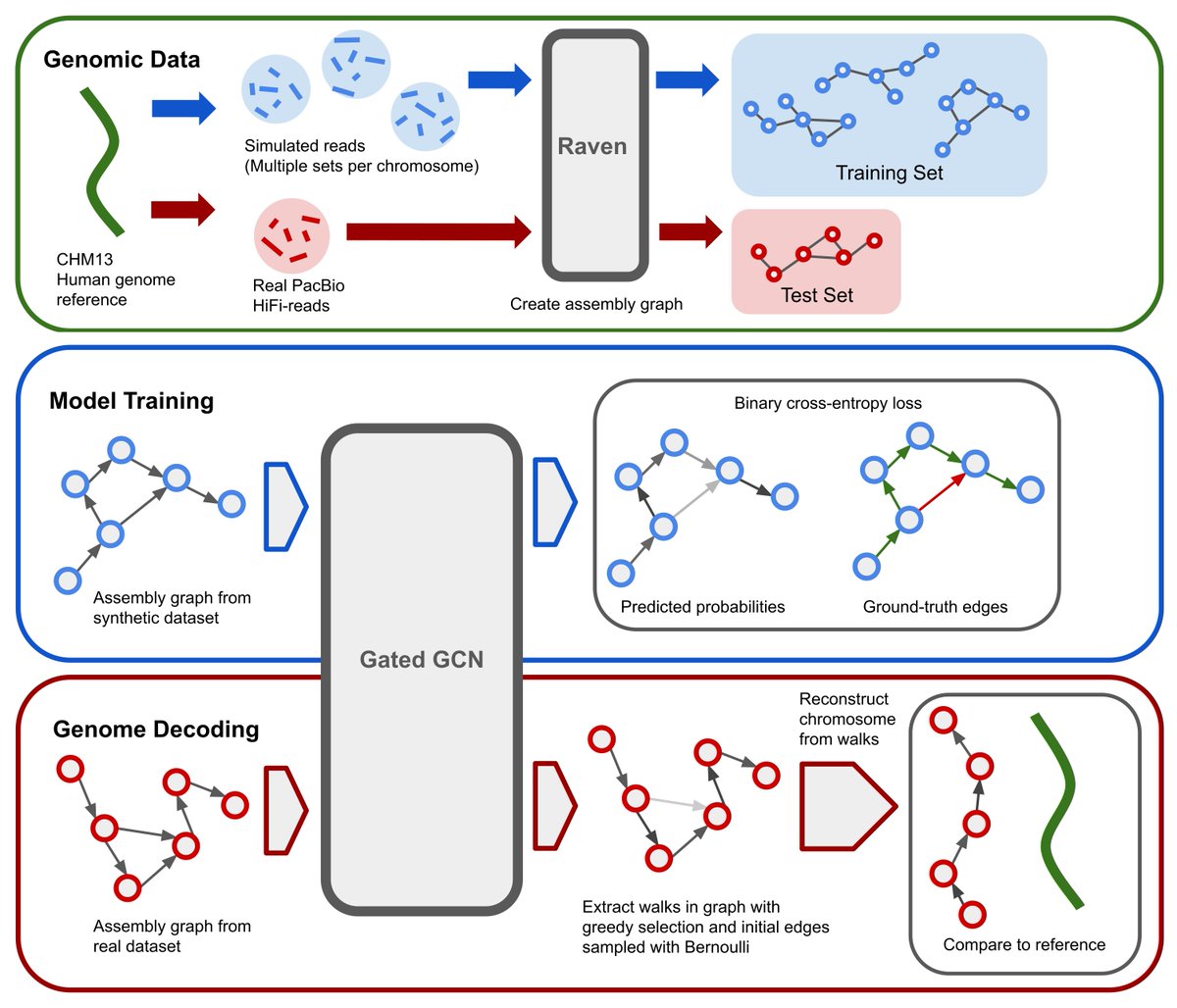
GNNome was published in @genomeresearch! This is a novel paradigm for de novo genome assembly based on GNNs. Without explicitly implementing any simplification strategies, it can achieve results comparable or higher than other SOTA tools. Paper, code, and overview are 👇 [1/8]
4
41
133
RT @KexinHuang5: 📢 Introducing Biomni - the first general-purpose biomedical AI agent. Biomni is built on the first unified environment fo….
0
117
0
RT @avapamini: How well do single-cell foundation models perform w/o finetuning?. Our work @GenomeBiology shows that in zero-shot settings,….
0
48
0
RT @chaitjo: Equivariance is dead - long live equivariance?. Scaling pure Transformers is the future of molecular generative models?. I thi….
0
16
0
RT @michael_galkin: 📣 Our spicy ICML 2025 position paper: “Graph Learning Will Lose Relevance Due To Poor Benchmarks”. Graph learning is le….
0
52
0
While the previous version of the tools was limited to graphs generated with the Raven assembler and was evaluated on individual human chromosomes, now it can take any OLC assembly graph as input and reconstruct genomes of entire organisms. [3/8].
After many months, I'm proud to share our work on untangling genome assembly graphs with GNNs. Or, GNNome Assembly (sorry not sorry)🧬. Paper: .Code/Data: . with @xbresson, T. Laurent, @Martin_fschmitz, and @msikic. Read more in 🧵
1
0
5
First, I would like to thank everyone without whom this work would not be possible: @msikic, @xbresson, @Martin_fschmitz, Thomas Laurent, and Kenji Kawaguchi. Paper: Code: 🧵 You can find the brief overview below! [2/8].
1
2
14
RT @msikic: Cannot agree more. Though-provoking. Recently, we showed the implementation of GNNs on genome assembly (huge graphs). https://t….
0
1
0
RT @ai4na_workshop: 🏆 We would once again like to thank our sponsors, who made it possible for us to provide you with lunch, and some of yo….
0
2
0
Organizing @ai4na_workshop was as challenging as it was rewarding. The room was full, the talks and posters interesting, and the panel engaging. This would not have been possible without @TinVlasic, @im50603 and @msikic. For the first-timers, I think we did a good job 😌.
Thank you all for attending AI4NA workshop! We hope you had a great time, learned something new, and made new connections. We would also like to hear from you–your feedback is valuable and we’ll do our best to take it into account for the future workshops. Link below 👇
1
1
9
I was very happy to see that AI conferences realized the world is bigger than the US, and what makes me happy now is how many people discovered how wonderful Singapore is :).
Singapore is the most impressive city I've ever visited. Way more than NYC or SF. I'm so happy that ICLR (& AABI) decided on Singapore, and I hope we can avoid the US for a bit 🙏 big thanks to my friends for showing me around and to all the awesome people I met and talked to 😊
1
1
6
RT @ai4na_workshop: Looking forward to seeing you tomorrow @iclr_conf ! Our workshop will start at 9 am and will take place in rooms Opal 1….
0
3
0