Michael Payne
@mjohnpayne
Followers
325
Following
342
Media
15
Statuses
114
PostDoc in @lanlabUNSW. Microbial genomics, Bioinformatics and Python!
Joined December 2014
Our new method for microbial genomic surveillance and epidemiology, multilevel genome typing (MGT) with the first database for Salmonella Typhimurium. Check out the paper https://t.co/N3RkEDJ0Mq and the new database https://t.co/KlnQL8FLqg.
#lanlab #Genomics #microbiology
2
11
23
Just published👍I’m glad to share: Genomic evidence of two-staged transmission of the early seventh cholera pandemic | Nature Communications @LanlabUnsw #cholera
nature.com
Nature Communications - The seventh cholera pandemic spread across the globe in three waves from 1961. Here, the authors sequence 22 genomes from 1961 to 1979 and show that the first wave of the...
0
6
9
New research from our talented PhD student Samitha! Genomic diversity of Salmonella Typhimurium isolates from meat chicken processing plants in NSW, Australia. #genomics #poultry #salmonella
frontiersin.org
Contamination of poultry products by Salmonella enterica serovar Typhimurium (STm) is a major cause of foodborne infections and outbreaks. This study aimed t...
0
2
3
Characterising Staphylococcus aureus genomic epidemiology with Multilevel Genome Typing. https://t.co/ojf2tkNRgL
#bioRxiv
0
2
1
New paper from @LanlabUnsw, in @EMI_Journal
https://t.co/ImYdwEXLYl. We described the global genomic epidemiology and evolution of B. #pertussis with >6500 isolates using MGT. @mjohnpayne @AliceZhengXu @compbiosan @sophie_octavia Public database -> https://t.co/czK2WACMzr. 1/6
1
5
9
Check out our latest work on “Genomic markers of drug resistance in Mycobacterium tuberculosis populations with minority variants”. Thank you all @drBenMarais @Sydney_ID
biorxiv.org
Minority variants of Mycobacterium tuberculosis harbouring mutations conferring resistance can become dominant populations during tuberculosis (TB) treatment, leading to treatment failure. Our...
0
3
12
New preprint from @LanlabUNSW! https://t.co/OilCkIUcDl We describe large and small scale trends in global #Bordetella #pertussis populations using a standardised multilevel genome typing (MGT) nomenclature and public database https://t.co/tlNrWJY1Il
@LanlabUnsw #microbialgenomics
1
5
19
Check out the collaborative work of @lanlabUNSW, Westmead hospital, and Queensland Health about #genomic epidemiology of Australian #Salmonella #Enteritidis using multilevel genome typing #MGT. Topics on #outbreak, #MDR and #plasmid. https://t.co/vBlpKCdmR9
1
3
6
My latest research on Genomic Epidemiology of Vibrio cholerae O139 is published on Emerging Infectious Diseases journal. Thank you @LanlabUnsw @mjohnpayne @DazhiJin @UNSWBABS #cholera is my favorite forever 😄
1
6
13
Thank you to @AUSSOCMIC for the ECR poster prize at #2022ASM. What a great meeting! Looking forward to next year! Congrats also to all the other members of @LanlabUnsw who contributed. @compbiosan, @AliceZhengXu, Dalong Hu and Ruiting Lan!
2
4
41
Come have a chat tonight at poster 112 at #2022ASM where I am presenting a new genomic nomenclature for Bordetella #pertussis using multilevel genome typing (MGT) developed in @LanlabUnsw. Also check out @compbiosan's poster 147 on our online database: https://t.co/KlnQL8FLqg!
0
2
11
Check my ePoster #209 of #2022ASM about plasmids contributing to the persistent colonisation of Listeria monocytogenes in the food markets @LanlabUnsw. It is a collaborative project with China CDC, and also an ongoing study of my previous research.
4
3
15
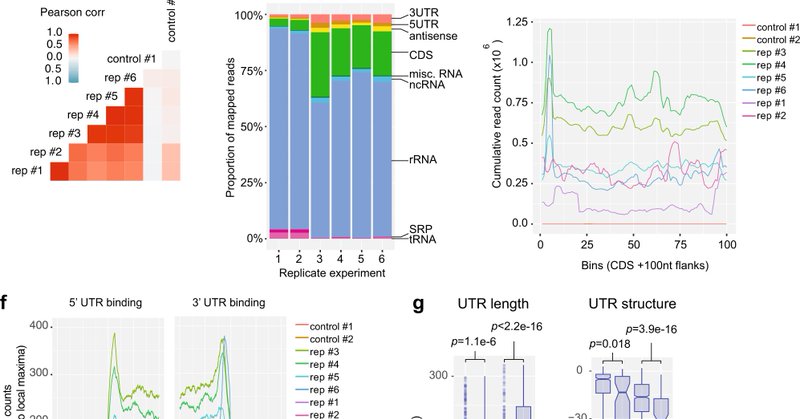
Super excited to share our latest work in the @jaitreeau lab published in @NatureComms detailing how a regulatory mRNA 3’UTR mediates vancomycin tolerance in a clinical vancomycin-intermediate Staph isolate. 🧵 1/3 https://t.co/wBksSGZF0u
nature.com
Nature Communications - Regulatory small RNA (sRNA) interact with mRNAs to regulate their stability, transcription, and translation via diverse mechanisms. Here, Mediati et al. apply RNase...
1
11
50
A summary 🧵of @LanlabUnsw latest Australian Bordetella pertussis genomics paper, just published in Emerging Microbes & Infection. Paper led by @AliceZhengXu who examined the microevolution of 385 B. pertussis strains across two epidemics from 2007-2018. https://t.co/8UGIJhdTjo
tandfonline.com
Whooping cough (pertussis) is a highly contagious respiratory disease caused by the bacterium Bordetella pertussis. Despite high vaccine coverage, pertussis has re-emerged in many countries includi...
1
6
15
Check out the great work from @hirokisuyama1 @LanlabUnsw His first ever pre-print. More info about the paper below 👇
New preprint! Integrating proteomic data with metabolic modelling provides insight into key pathways of Bordetella pertussis biofilms
1
4
13
I am going to give a seminar talk at University of Technology Sydney about #genomic #epidemiology of #Salmonella Enteritidis using a global database MGTdb @LanlabUnsw Please join us online on 27th April (this Wed) at 12:30 p.m. AEST. zoom link: https://t.co/DeZRWcMhOX
3
4
24
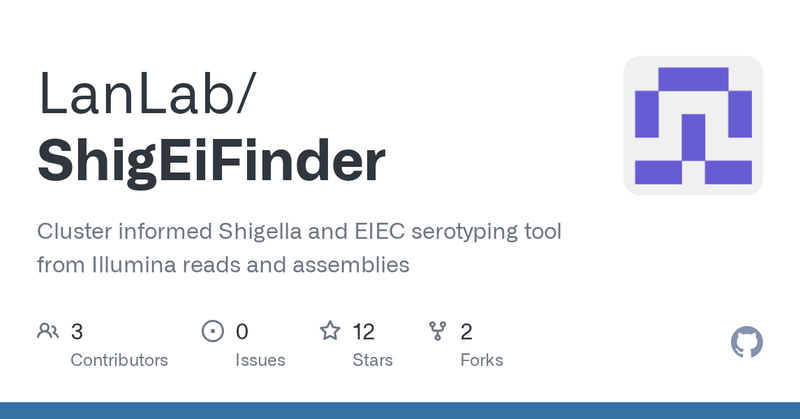
ShigEiFinder version 1.3 released! Includes fixes and updates that resolve issues raised in this excellent recent paper by @iman__yassine and @weill_xavier
https://t.co/JFM4Lt9IEV. Accuracy now over 92% in routine surveillance! @LanlabUnsw #Shigella #EIEC.
github.com
Cluster informed Shigella and EIEC serotyping tool from Illumina reads and assemblies - LanLab/ShigEiFinder
0
5
8
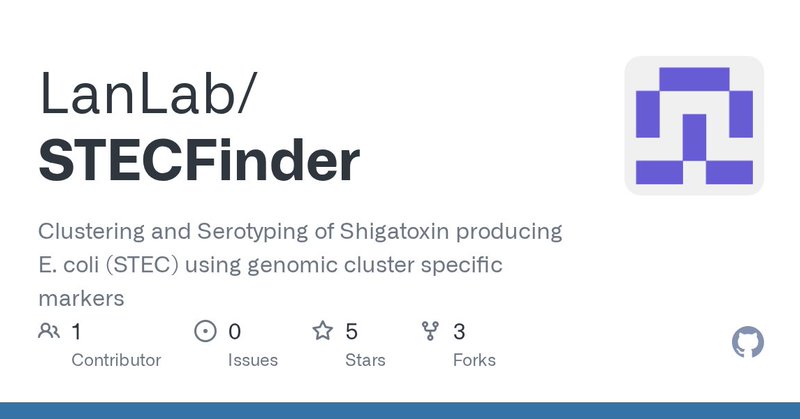
New version of the Shiga toxin-producing E. coli genomic clustering and serotyping tool, STECFinder (v1.1)! Allows detection of multiple stx2 types in the same strain amongst other improvements. Available on github and conda https://t.co/Mn3BgwBY5S.
@LanlabUnsw #MicrobialGenomics
github.com
Clustering and Serotyping of Shigatoxin producing E. coli (STEC) using genomic cluster specific markers - LanLab/STECFinder
0
3
8
Thrilled to be at @GenomeConf for my first in-person conference in >2 years & excited to be presenting my poster on a novel regulatory mRNA-mRNA interaction in the prokaryote S. aureus.
0
4
42