Jon Belyeu
@jon_belyeu
Followers
314
Following
178
Media
18
Statuses
149
Bioinformatics scientist at PacBio. Views expressed are my own and not those of employer, etc.
Salt Lake City, UT
Joined September 2015
I'm starting to transition away from here so I wrote a longer summary of the manuscript where the skies are blue 🦋. If you aren't on there yet, there's no time like the present 😀
0
0
0
I just released a new preprint! The manuscript describes SVTopo, a software tool that enhances visualization of complex SVs using HiFi data:
biorxiv.org
Structural variants are genomic variants that impact at least 50 nucleotides and can play major roles in diversity and human health. Many structural variants are complex multi-breakpoint rearrangem...
1
0
4
“StarPhase: Comprehensive Phase-Aware Pharmacogenomic Diplotyper for Long-Read Sequencing Data” is now available online! In this work, we explore the use of long-read sequencing (#PacBio #HiFi) for #pharmacogenomics #PGx. 1/2
1
3
11
It's official, @PacBio has launched a new benchtop sequencer! Quick summary of the new Vega system: - instrument = $169k - consumables = $1100 per run - output = 60 Gbp, 24 hr run time This is the #HiFi sequencer #microbiology labs have been asking for. https://t.co/RYJdUCq8SA
pacb.com
Priced at just $169,000 Vega is designed to make highly accurate long-read sequencing accessible to any laboratory Translations: Japanese | Chinese | Korean MENLO PARK, Calif., Nov. 06, 2024...
4
37
111
Check out these examples (from HG002):
github.com
Complex structural variant visualization for HiFi sequencing data - PacificBiosciences/SVTopo
0
0
3
HiFi sequencing does a great job of identifying structural variation, but I've spent many hours struggling with visualization tools to understand what's really going on in complex rearrangement
1
0
1
I'm happy to announce that SVTopo, a new computational tool from PacBio ( https://t.co/W6sHuB0Ps9), is ready for you to try. SVTopo creates simple images so you can rapidly understand complex changes in genome structure. See me and learn more at ASHG 2024 in Denver, poster 1180W
github.com
Complex structural variant visualization for HiFi sequencing data - PacificBiosciences/SVTopo
2
24
60
Happy to introduce sawfish, a new HiFi structural variant caller emphasizing local haplotype modeling to improve SV representation and genotyping in both single and joint-sample analysis contexts: 1/n https://t.co/P3kp1QUd45
biorxiv.org
Motivation Structural variants (SVs) play an important role in evolutionary and functional genomics but are challenging to characterize. High-accuracy, long-read sequencing can substantially improve...
2
21
64
A familial, telomere-to-telomere reference for human de novo mutation and recombination from a four-generation pedigree https://t.co/vOAv3jhRcT
#biorxiv_genomic
biorxiv.org
Using five complementary short- and long-read sequencing technologies, we phased and assembled >95% of each diploid human genome in a four-generation, 28-member family (CEPH 1463) allowing us to...
0
24
36
I’m happy to say that we’ve just released a manuscript on bioRxiv describing our new tool TRGT-denovo. This new method was developed by Tom Mokveld and @egor_dolzhenko along with the help of many collaborators (1/n)
biorxiv.org
Motivation Identifying de novo tandem repeat (TR) mutations on a genome-wide scale is essential for understanding genetic variability and its implications in rare diseases. While PacBio HiFi sequen...
1
21
47
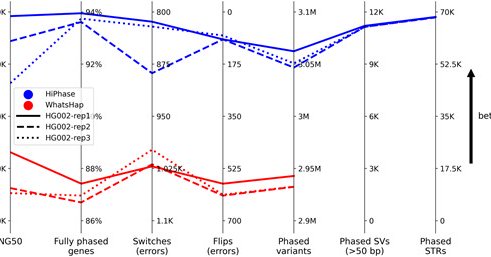
Excited to announce that our accompanying paper on HiPhase has been published today, I'll highlight some of the additions in this thread! #PacBio #HiFi “HiPhase: Jointly phasing small, structural, and tandem repeat variants from HiFi sequencing”:
academic.oup.com
AbstractMotivation. In diploid organisms, phasing is the problem of assigning the alleles at heterozygous variants to one of two haplotypes. Reads from Pac
2
20
44
Characterization and visualization of tandem repeats at genome scale https://t.co/zSE5haiB3r
1
54
175
Great @biorxivpreprint: how you can call LPA CNV from @illumina WGS. This is a sig. step forward for studying #cardiovascular risk. 60% of samples report phased CNV -> improves phenotype correlation. @BCM_HGSC @MikeEberle @jon_belyeu @srbehera11
https://t.co/03aedlT7bR
1
13
30
Checkout PB2257 #ASHG22 by #Illumina's @jon_belyeu, @sedlazeck and Baylor U. They show how to estimate total and allele-specific KIV-2 repeat length, which strongly impacts #CVD risk. A challenging genomic region that provides enormous medical insight, coming to #DRAGEN soon.
0
6
16
Very happy to share my first research article @illumina on genotyping HBA1 and HBA2 genes from whole-genome sequencing data: https://t.co/4F87S1V6ku.
illumina.com
4
9
35