Mike Eberle
@MikeEberle
Followers
1K
Following
8K
Media
53
Statuses
2K
Husband | Father | Scientist @PacBio
San Diego, California
Joined October 2011
Happy that our manuscript describing TRGT is now available. If you would like to learn more about the importance of tandem repeats and the variation that must be measured when studying them, please also check out the accompanying research briefing:
3
37
109
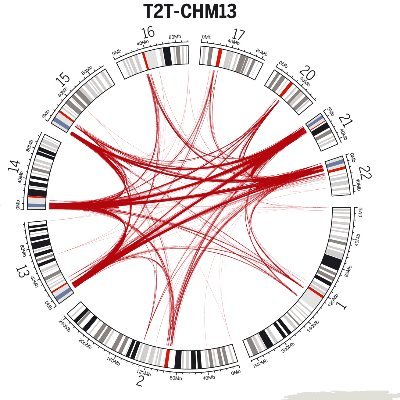
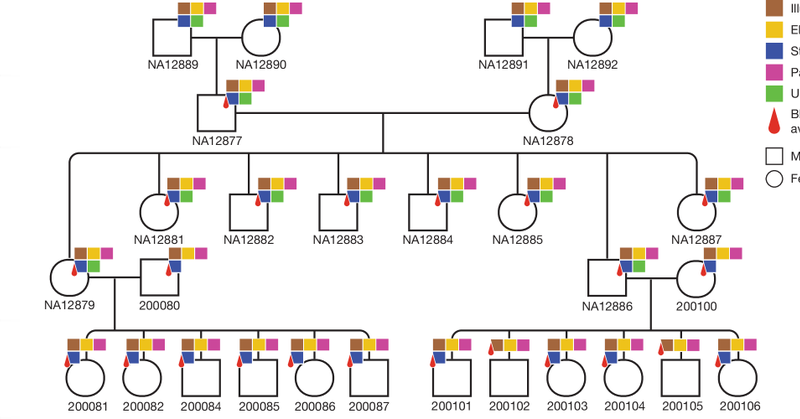
Our Nature paper ( https://t.co/8ucXysP2w8) deep sequencing a 4-generation, 28-member family using multiple sequencing technologies to study transmission of all classes of genetic variation is out! @EichlerEE @uwgenome @HHMINEWS
Advanced genomic analysis of 4 generation family offers new knowledge about genetic mutations & their transmission, including inherited variants & those that arise anew @Nature @uwgenome @EichlerLab @UWMedicine @UUtah @PacBio
https://t.co/DRYYOenRZm
2
23
61
Advanced genomic analysis of 4 generation family offers new knowledge about genetic mutations & their transmission, including inherited variants & those that arise anew @Nature @uwgenome @EichlerLab @UWMedicine @UUtah @PacBio
https://t.co/DRYYOenRZm
0
2
9
A first in human genomics: a 4-generation pedigree reference, now in Nature. Built with #PacBio HiFi, it maps de novo mutation rates, reveals paternal bias, and captures high mutation rates in tandem repeats—even in Y and repeat-rich regions. Paper here:
nature.com
Nature - Analysis of more than 95% of each diploid human genome of a four-generation, twenty-eight-member family using five complementary short-read and long-read sequencing technologies provides a...
Advanced genomic analysis of 4 generation family offers new knowledge about genetic mutations & their transmission, including inherited variants & those that arise anew @Nature @uwgenome @EichlerLab @UWMedicine @UUtah @PacBio
https://t.co/DRYYOenRZm
1
3
6
Nature research paper: Human de novo mutation rates from a four-generation pedigree reference https://t.co/2IlOKYGjIs
nature.com
Nature - Analysis of more than 95% of each diploid human genome of a four-generation, twenty-eight-member family using five complementary short-read and long-read sequencing technologies provides a...
8
55
191
“StarPhase: Comprehensive Phase-Aware Pharmacogenomic Diplotyper for Long-Read Sequencing Data” is now available online! In this work, we explore the use of long-read sequencing (#PacBio #HiFi) for #pharmacogenomics #PGx. 1/2
1
3
11
I will be hosting an information session for applicants to the USC Computational Biology and Bioinformatics PhD program, discussing research topics and application strategies. If you are registering with a non-institutional e-mail, please let me know https://t.co/mw1NX3Fof3
0
10
11
Just back from #ashg24 - new record by an automated library prep for a Revio SMRT cell: 8.1 M reads; 133.0 Gb; 16.4 kb avg size; Q34. …and this is before upgrade to SPRQ! Great lab team @GeneticNijmegen @radboudumc! #RevioOlympics
0
5
17
Fantastic turnout at #ASHG24 for Xiao Chen’s platform talk! 🎉 She shared exciting insights on using #HiFisequencing for genome-wide profiling of highly similar paralogous genes. Thank you to everyone who joined us to explore the power of #PacBio technology! #ASHG2024
0
2
11
Great talk by Xiao Chen about application of Paraphase in Four Seasons 4. Learn more about the method in last year’s AJHG paper #ASHG24
https://t.co/4ZvZwNGBK4
cell.com
We developed Paraphase, an informatics method that, combined with highly accurate long reads, can resolve the highly homologous SMN1/SMN2 genes involved in spinal muscular atrophy. We characterized...
0
8
19
Xiao talking now about using Paraphase and HiFi data to resolve segmental duplications genome wide
0
3
21
Was your answer also pharmacogenomics? Then I have some exciting things to show you at #ASHG24... Looking forward to next week in Denver! #PacBio #PGx
The excitement around #ASHG24 is building, and we’re just getting started. We recently asked our community to share their vision for the future of #genomics. Now, watch this special video featuring some of your responses recorded by the #PacBio team.
0
1
8
Excited to be at #LRUA24 Long-reads meeting in Uppsala where I get to meet so many friends and learn about amazing science and technology @LongtrecEU will be leading the lrRNAseq workshop!! @PacBio @nanopore
0
2
25
Are you interested in targeted #sequencing? Make sure to check out Tom Mokveld's #PacBio poster at #LRUA24 to learn more!
0
4
17
Join us at #ASHG24 for a featured symposium from @egor_dolzhenko on profiling genetic and epigenetic variation in tandem repeat regions using #HiFisequencing. Learn how this approach is advancing precision #genomics research. Details: https://t.co/3ZPGVoXPec
#PacBio
0
6
12
What a stage! ✨ @Somar1987 presenting our #PacBio intro talk this morning at #LRUA24. If you have any follow-up questions, make sure to stop by our booth. #HiFiSequencing
0
6
14
Help shape MND/ALS research! 🧠 LifeArc’s new survey focuses on tech to improve daily life with MND. Last time, @lifearc1 received 200 responses—thank you! 🙏 📢 If you have MND/ALS, care for someone, or are at risk, your input is crucial. @lifearc1
What technology could help improve daily life for those living with #MND? We'd like to better understand the daily challenges to guide the development of new technologies and inform our future research and investment. Share your thoughts in our survey: https://t.co/0ntuTIyU1b
0
7
18
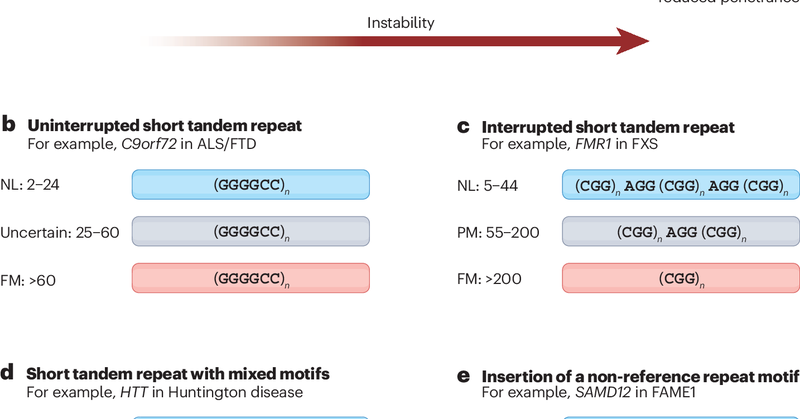
Another #Review highlight: Sequence composition changes in short tandem repeats: heterogeneity, detection, mechanisms and clinical implications https://t.co/PZjcoh7M6k by @IndhuShreeRB, @egor_dolzhenko, @MikeEberle & Jan M. Friedman @UBC @BCCHresearch @PacBio
nature.com
Nature Reviews Genetics - This Review highlights the diversity in sequence composition of disease-related short tandem repeats. The authors discuss how to detect non-canonical motifs in repeat...
1
3
15