Hongkai Ji
@jihk99
Followers
794
Following
31
Media
0
Statuses
42
Professor @jhubiostat, data scientist
Joined September 2014
Exciting collaboration with @Dr_CJackson, @dpardol1, @BettegowdaMDPHD, @Yegnalab, and many others - now published in Science! Grateful for the contributions of our current and past group members: @stephlee51, @ZhichengJi, @HWenpin, and Wentao Zhan. #TeamScience.
New research from @Dr_CJackson (@PennNSG) out in @ScienceMagazine identifies 14 distinct immune-suppressing cell subsets in #glioblastoma tumors, shedding light on how the tumors evade the immune system & spread ft. Kate Jones
0
3
7
FUNCODE scores are now available on the ENCODE Data Portal: and UCSC Genome Browser: Huge thanks to @ENCODE_NIH, @GenomeBrowser, @WeixiangFang, and our amazing team for making this possible! 🙌🧬.
0
0
1
Big thanks to @GenomeBrowser for integrating FUNCODE into the UCSC Genome Browser!.
We are proud to announce our latest Public Hub, FUNCODE. FUNCODE (Functional Conservation of DNA Elements) is a computational framework that quantifies functional conservation between human/mouse regulatory elements using ENCODE functional genomics data.
0
0
1
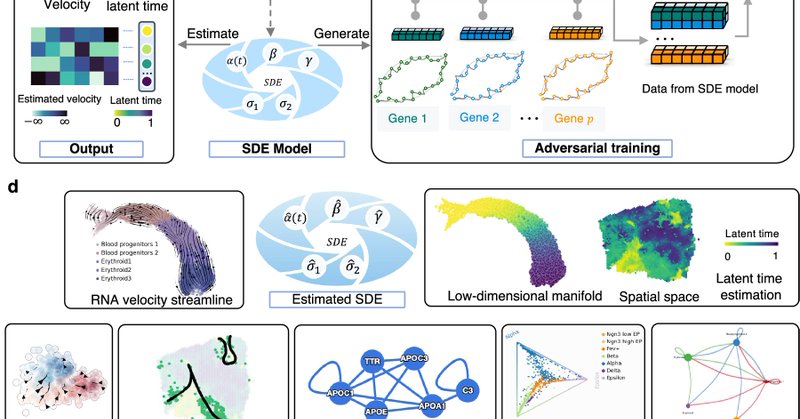
SDEvelo, a generative approach for inferring RNA velocity using multivariate stochastic differential equations (SDE). @gordonliu810822.
nature.com
Nature Communications - RNA velocity predicts directed cell trajectories but often fails to capture inherent randomness, leading to false positives from random patterns. Here, authors present...
1
0
5
Excited to share Lamian - a statistical framework for differential pseudotemporal trajectory analysis across biological conditions using single-cell RNA-seq data from multiple samples. @HWenpin @ZhichengJi @ZeyuCHEN19 @EJohnWherry @stephaniehicks .
3
6
52
This is jointly with the wonderful @kalhorlab & Zack lab. It is part of @WeixiangFang & Claire Bell's thesis work,.
1
0
4
Glad to share quantitative fate mapping (QFM), now officially online @CellCellPress. QFM uses cells' lineage barcodes to retrospectively infer and quantify the stochastic process of cell fate determination.
3
9
56
RT @LabVokes: Our study showing that Hedgehog signaling is essential for maintaining the early foregut and larynx by preventing EMT was pub….
elifesciences.org
HH signaling prevents precocious EMT and cell death to maintain epithelial cell states during early stages of larynx development.
0
6
0
RT @SmithImmunology: #vaccination diversifies the #SARSCoV2-specific T cell repertoire in patients with a prior history of #COVID. Shout o….
0
1
0
RT @kalhorlab: Lineage barcoding approaches were limited to measuring static features of development. Preprints from us & @TanjaStadler_CH….
0
13
0
It is my honor to be appointed as the co-Editor-in-Chief of Statistics in Biosciences. Look forward to working with our authors, associate editors, and EIC Dr. Joan Hu to make SIBS a great platform for disseminating your impactful work and inspiring transformative research.
10
2
82
Trick or treat? For this Halloween, our treat for those who are analyzing single-cell RNA-seq data is - TreeCorTreat.
biorxiv.org
Single-cell RNA-seq experiments with multiple samples are increasingly used to discover cell types and their molecular features that may influence samples’ phenotype (e.g. disease). However, analyz...
3
9
59