Dan Hebert
@hebERt_lab
Followers
157
Following
634
Media
0
Statuses
20
ER cell biologist, glycoprotein homeostasis, @UMassBMB, @UMassMCB
Massachusetts, USA
Joined December 2018
RT @chienlab: We are looking for a new colleague to join my department, the focus is on quantitative or structural biology, let me know if….
0
10
0
Thank you Marcin!.
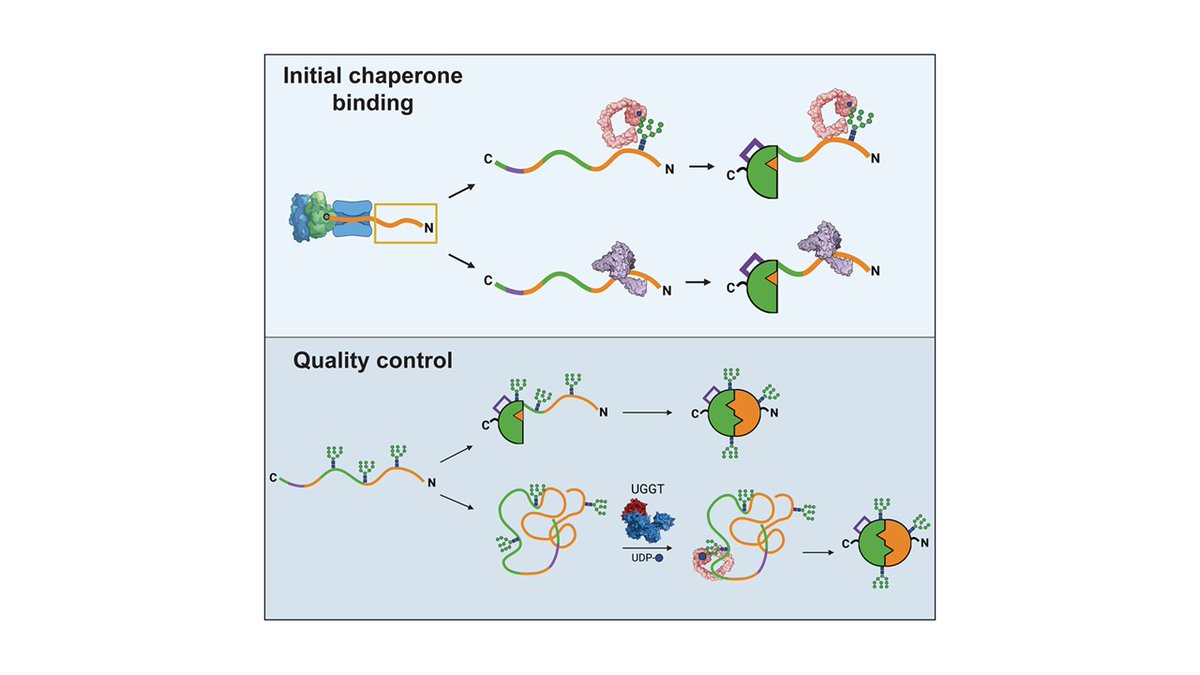
Elegant study shows how the location and composition of N-glycans (glyco-code) influences protein folding and quality control. Competition between early lectin chaperone and BiP, and the role of UGGT particularly ineteresting. @hebERt_lab #glycotime
0
1
7
RT @pietro1968: A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein folding quality control checkpoint: iScience https://….
0
3
0
And co- and post-translational modifications like glycosylation.
@MrSteveKnutson There is no "right" answer (and some of the Ub OGs bounce around in their papers), but there IS a right answer:. Ubiquitylation. It matches the nomenclature rules of other PTMs (methylation, phosphorylation, acetylation, etc.).
0
0
4
RT @chienlab: If you have any questions, feel free to check out the ad or get in touch! [can you see yourself here?⬇️] .
0
1
0
RT @TobiWalther: This fall the @FandWLab is moving to the #cellbiology department @MSKCancerCenter. Interested in fundamental, mechani….
0
43
0
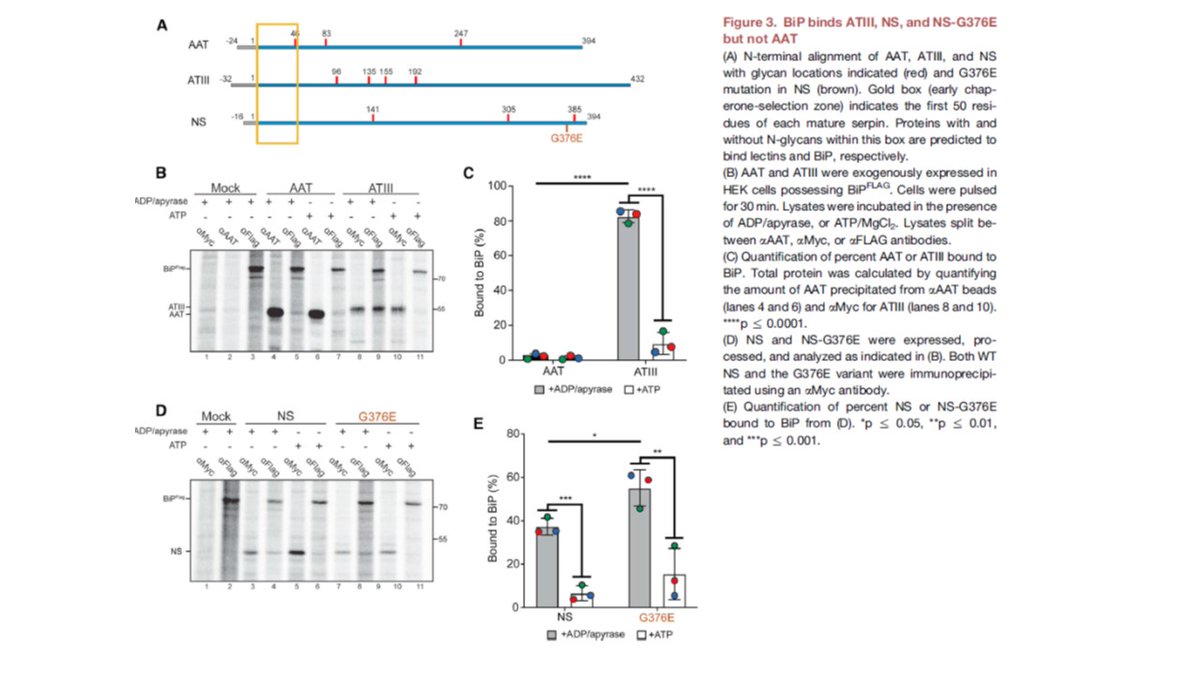
This is a long-term collaborative study with @gierasch1 and Anne Gershenson from @UMassAmherst and Terry Flotte of #UMassChan that builds off an initial study using SAHA by Balch and colleagues. #ER_literature #glycotime.
1
0
1
Congrats to Haiping Ke and @kpguay28 for their work that shows that a small molecule proteostasis regulatory boosts muscle cell alpha-one secretion, the site of intramuscular injections for gene therapy.
1
1
7
Excited to share our new collaborative preprint from @pietro1968, @zitzmannlab, @kpguay28 and others that describes the discovery of the first inhibitor to the ER quality control sensor UGGT. Something for everyone: crystallography, NMR and cell biology studies! #glycotime.
A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein folding quality control checkpoint
1
8
25
RT @theNASciences: Elected in 2019, Lila M. Gierasch of @UMassAmherst @UMassBMB is being inducted to the National Academy of Sciences for i….
0
7
0
How ER UGGT recognizes non-native substrates remains a mystery. This exciting collaborative study led by @pietro1968 reports on the structure of UGGT and uses MD to model molecular motions of its 4 thioredoxin domains to provide valuable insight into its interrogation methods.
Pietro Roversi @pietro1968 and colleagues at @LeicStructBio describe the inter-domain motions in the #misfold-recognising portion of #UDP-glucose:#glycoprotein glucosyl-transferase #StructureCP
1
1
10
Thanks Mauri.
We highlighted for F1000 a nice ELife paper on protein quality control in the mammalian ER. Great job by Dan Hebert's guys! UGGT1 and UGGT2 ensure the quality of our proteome. Quantitative glycoproteomics reveals . - recommended in @FacultyOpinions
0
0
5
Congrats to grad student @_benjamin_adams for developing a glycoproteomics platform to ID natural substrates of key quality control ER folding sensors UGGT1 and UGGT2 and revealing UGGT2’s preference for lysosomal proteins. Good luck on his next endeavor @sshaolab @HarvardCellBio.
Happy to have our work identifying cellular substrates of the ER folding sensors UGGT1 and UGGT2 in @eLife. @UMassMCB @hebERt_lab.
0
1
9