Feng Alex Yan
@alexyfyf
Followers
330
Following
2K
Media
65
Statuses
974
#bioinformatics #cancer
Melbourne, Victoria
Joined March 2015
🚀 Prophet v3 is out! AI that predicts how cells respond to genetic & chemical perturbations across readouts. Now scales to 1.9 M molecules + in vitro validation of melanoma-specific hits 🧬💊 📄 https://t.co/JTd0VQUXls
#AI #Biology #DeepLearning
4
51
196
Not long now until our Long-Read Research Symposium on November 13th. See below the amazing line-up of speakers and talks. It's not too late to register for this free event - click the link --> https://t.co/vMc0x3mwmz
0
2
3
Pre-phasing long reads improves structural variant genotyping https://t.co/aHCO8nDTCc 🧬🖥️🧪 https://t.co/PqrDE6Xv3o
1
12
30
Longdust, a new tool to identify highly repetitive STRs, VNTRs, satellite DNA and other low-complexity regions (LCRs). Similar to SDUST but for long regions. https://t.co/xUFgpmatJ5
github.com
Identify long STRs, VNTRs, satellite DNA and other low-complexity regions in a genome - lh3/longdust
2
70
201
While everyone was obsessing over CRISPR, a small team just quietly published a paper in Science solving genetic medicine's biggest problem. They created a system that can fix thousands of different mutations at once. Here's how they did it 🧵
45
582
3K
🔬Applications for the EMBL Australia #PhD Course are now OPEN!🌟 Ready to kick-start your scientific career? Don't miss this opportunity to learn from Aus's best researchers! 📅 1-12 December, 2025 📍 @SAiGENCI, Adelaide ℹ️ Info & apply by 27 July: https://t.co/BdhInFxbLi
1
19
25
Ultrafast and accurate sequence alignment and clustering of viral genomes https://t.co/0SBYk0zFYN 🧬🖥️🧪 vclust https://t.co/ckPsMo8LY4 clusty https://t.co/TBpd4feY5z
0
8
32
Review: Transcriptomics in the era of long-read sequencing https://t.co/HeYItvnv0t (read free: https://t.co/cLJHVSZqWL) 🧬🖥️🧪
0
33
116
Part 1 and 2 of the Long-read sequencing Special Issues are now live! https://t.co/izr399i5mj
0
34
99
scnanoseq https://t.co/zdjMcNv6nS: an nf-core [Nextflow] pipeline for Oxford Nanopore single-cell RNA-sequencing https://t.co/dNcKi4dcFe 🧬🖥️🧪 https://t.co/sqX0vVovpu
1
17
71
📣📣2 weeks until Abstract Deadline!!! Get your abstracts in now - you don't want to miss out.
📢 Registrations and Abstract submissions are now open for Cancer Bioinformatics Australia 2025! Abstracts: 11th April https://t.co/aemSgmbUlJ Registration: https://t.co/o4MRpikHpP Further info: https://t.co/RjsabOsoL0
0
2
3
Our new paper examining how to analyse longread RNA-seq with no reference genome. We compare approaches for assembly and downstream analysis, from transcript accuracy to differential expression. Lead by @alexyfyf. Thnx to all contributors incl. @QGouil for the pea data!
Towards accurate, reference-free differential expression: A comprehensive evaluation of long-read de novo transcriptome assembly https://t.co/CR7YjdjTRW
#biorxiv_bioinfo
0
11
28
I am looking for a postdoc to join my new team in Melbourne, Australia at WEHI to work on lung stem cells and disease, including lung cancers! International candidates+ those just finishing up PhDs welcomed Please apply below: https://t.co/VUVq7XwoMf
4
87
177
For anyone who has struggled with code navigation of the nested @nf_core shared modules, rejoice! The new @nextflowio #VSCode extension code navigation makes this super fast! 🎉
Powerful code navigation features let you (alt/ctrl) + click on imports, processes and workflows to go directly to their definition. This means you no longer need to dig through the file navigation to see where the code is being defined!
0
10
20
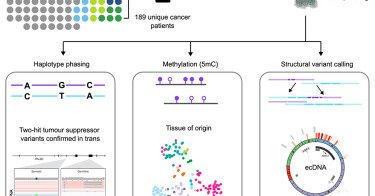
#WGS long-read sequencing of #cancer using @Nanopore adds yield - particularly resolution of structural variation, identification of #HRD, #phasing and discovery of allele-specific #methylation of tumor suppressors #genomics
https://t.co/NzExp1Jgxf
cell.com
We present a resource dataset of 189 long-read-sequenced tumors from advanced cancer patients. Long-read sequencing allows detection of a range of features not easily detectable with more conventio...
2
20
59
𝗦𝗧𝗔𝗠𝗣: single cell (spatial?) Being asked what I think about it (NOT involved). 1) What is STAMP? Should I care about it? STAMP is a clever way of using 𝘀𝗽𝗮𝘁𝗶𝗮𝗹 𝗮𝘀𝘀𝗮𝘆𝘀 to get 𝗶𝗻𝘀𝗮𝗻𝗲𝗹𝘆 𝗹𝗼𝘄 𝗰𝗼𝘀𝘁 𝘀𝗶𝗻𝗴𝗹𝗲 𝗰𝗲𝗹𝗹 𝗱𝗮𝘁𝗮 https://t.co/v9nNsbJ5kx
2
23
103
Google has a new experimental tool (Illuminate) that takes a link to a preprint and creates a "podcast" discussing the paper. I tested this with a few preprints that recently caught my attention (Ropebwt3, genomic language models, VEP guidelines)
blog.stephenturner.us
Listen now | Google's new Illuminate tool takes a PDF from arXiv and generates a short podcast discussion on the paper's key points
2
36
136
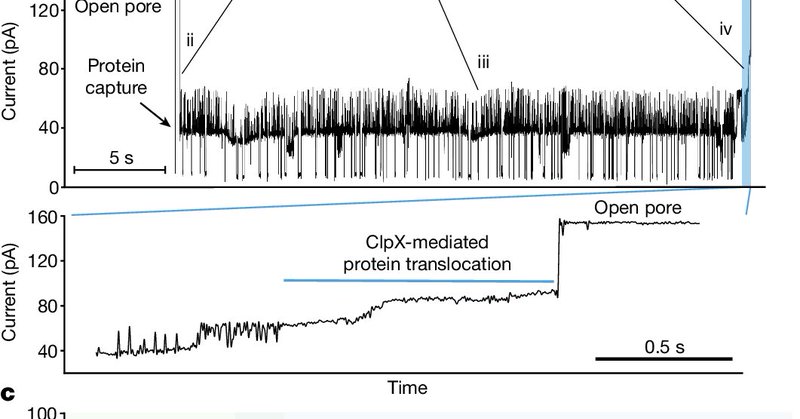
Published today in @Nature, we describe an approach for single-molecule protein reading on @nanopore arrays. By utilizing ClpX unfoldase to ratchet proteins through a CsgG nanopore, we achieved single-amino-acid sensitivity.
nature.com
Nature - A technique for threading long protein strands through a nanopore by electrophoresis and back using a protein unfoldase motor, ClpX, enables single protein molecules to be analyzed...
19
287
997
📢 Registrations for the "Hello Nextflow!" workshop close tonight!
biocommons.org.au
Get started on the path to writing your own reproducible and scalable scientific workflows using Nextflow. More information Apply here
0
3
3