Genomic Technologies Group
@GenTechGp
Followers
663
Following
232
Media
15
Statuses
287
Genomic Technologies Group // The Garvan Institute of Medical Research // Australia's leading long-read sequencing service // Enquiries: [email protected]
Joined August 2020
Cornetto v0.2.0 is now released for using programmable selective nanopore sequencing for genome assembly. GitHub: https://t.co/Sp2CHp1BmT Paper: https://t.co/9y00QcmBfF Datasets: https://t.co/8cFKDUcoZh ... and a banner made by Ira @GenTechGp referring to ‘no boring bits’.
Our cornetto work is now published at https://t.co/9y00QcmBfF Cornetto can do near-T2T assembly using @nanopore adaptive sampling (readfish by @mattloose) & hifiasm by @ChengChhy - with less 💸 - reference agnostic, so works for non-humans - not just blood, even saliva
1
14
40
Not long now until our Long-Read Research Symposium on November 13th. See below the amazing line-up of speakers and talks. It's not too late to register for this free event - click the link --> https://t.co/vMc0x3mwmz
0
2
3
Join us for a day (Nov 13) of fascinating talks and discussions on the latest in long-read sequencing tech and related research in genomics, transcriptomics, and epigenetics. A great opportunity to network and hear from leading experts from around Aus. https://t.co/DLugyQwJ6u
eventbrite.com.au
Join us at the Garvan Long-Read Research Symposium 2025 for a deep dive into groundbreaking discoveries in medical research!
0
5
12
Tha Garvan long read research symposium is BACK! Sponsored by @nanopore and @PacBio this event is FREE to join. Learn all about the fun you can have with long reads 🤓 register below https://t.co/TynhVx5YgO
eventbrite.com.au
Join us at the Garvan Long-Read Research Symposium 2025 for a deep dive into groundbreaking discoveries in medical research!
1
3
8
Come join us this November 13th for the Garvan Long-Read Research Symposium! You'll hear about the wonderful things you can do with @nanopore and @PacBio long reads from a great line-up of speakers. FREE to attend. Register below https://t.co/vMc0x3mwmz
eventbrite.com.au
Join us at the Garvan Long-Read Research Symposium 2025 for a deep dive into groundbreaking discoveries in medical research!
0
6
8
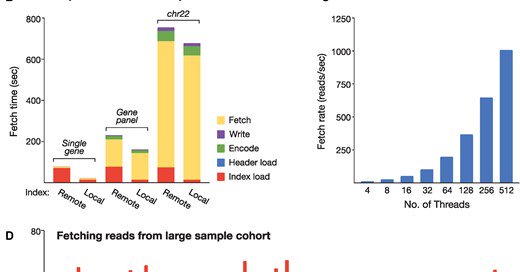
Check out our detailed evaluation of SLOW5 vs POD5 format for raw Nanopore data. TLDR: SLOW5 still the best nanopore data format.
For many of those who were asking on BLOW5 vs POD5 for nanopore signal data, here is a finally detailed benchmark we did: https://t.co/ZspXSlrW9e Summary: performance of BLOW5 is >= POD5 (from ~= to 100X, see below), with benefit of having ~3 dependencies instead of >50.
0
3
6
Check out the excellent line-up of speakers we have for the @GarvanInstitute Long Read Research Symposium day It is FREE to come along - this November 7th. Reserve your spot https://t.co/fvgVtzOpfs
0
2
2
Not long now until the @GarvanInstitute Long Read Research Symposium on November 7th. We will have talented speakers covering the advantages of both @nanopore and @PacBio data Only a few spots left - don't miss out! 💃 https://t.co/fvgVtzOpfs
0
6
9
Cut your Nanopore raw data files in half with our new compression method ex-zd. Thanks @Hasindu2008 for leading this project 🧬
Introducing ex-zd, a lossless+lossy signal compression for @nanopore signal data. While lossless can only save about 1-3% over vbz, lossy can cut the file sizes by almost half with no noticeable impact on basecalling or methylation calling accuracy. https://t.co/xnGcLOlaCQ
0
3
4
We are hosting a Long Read Research Symposium at @GarvanInstitute ! Our talented speakers will cover the advantages of both @nanopore and @PacBio data. Join us to find out what long reads could do for you 🧬 https://t.co/fvgVtzOpfs
0
15
22
Our #slow5curl paper is out! https://t.co/TbmcA8c1X4 Big projects storing hundreds/thousands of @nanopore signal datasets on cloud storage like @awscloud #s3 will be able to save 💸,⏲️ and bandwidth while improving the accessibility of datasets to those with limited computing ..
academic.oup.com
AbstractBackground. As adoption of nanopore sequencing technology continues to advance, the need to maintain large volumes of raw current signal data for r
2
17
50
Excited to share our latest work on somatic mutations in celiac disease! We discover expanded T cell clones with somatic driver mutations in individuals that don't respond to a gluten-free diet. With @FabioItaus, Chris Goodow and many others! Check it out:
medrxiv.org
Intestinal inflammation continues in a subset of celiac disease (CD) patients despite a gluten-free diet. Here, by applying multiomic single cell analysis to duodenal biopsies, we find low-grade...
0
10
38
Super excited to share our work on #celiac disease. We discovered T cells carrying somatic mutations which may explain chronic autoimmune disease https://t.co/UaZgVN76gW . Great team work led by @Manu___Singh. Thank you to all authors!
medrxiv.org
Intestinal inflammation continues in a subset of celiac disease (CD) patients despite a gluten-free diet. Here, by applying multiomic single cell analysis to duodenal biopsies, we find low-grade...
4
23
80
Want an easy to use and fast error/SNP tolerate grep-like tool? Have complex barcodes or indices to demultiplex from raw `omics data? Flexiplex is now published, 🥳 https://t.co/7oMnclGAQN, with some great software updates for even more flexibility, https://t.co/sgBXA7a3bQ 1/3
Pleased to report the very first paper from my research group at WEHI is now up on bioRxiv. Flexiplex is a new tool for raw sequencing data that allowed you to identify and error correct barcodes, and can even be used as a general "grep-like" search tool.
4
25
71
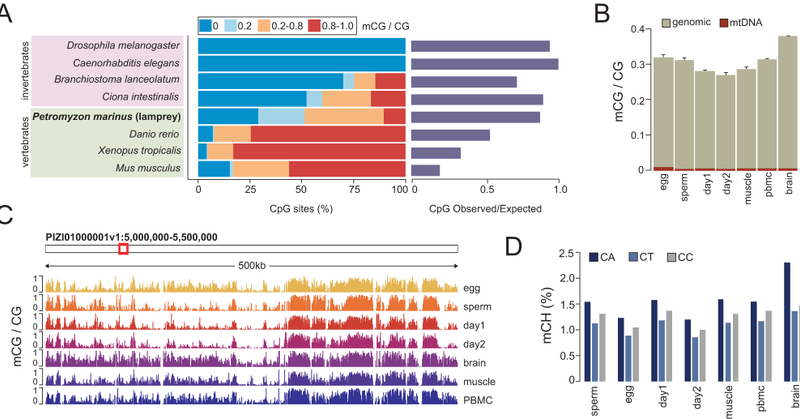
Final version of our lamprey epigenome remodeling paper is now out in @NatureComms
nature.com
Nature Communications - DNA methylation plays a major role in establishing cell identity, but the dynamics of DNA methylation patterns are highly variable across species. Here, the authors discover...
16
58
198
Squigualiser preprint with a bunch of usecases is now available 🔥. Your feedback is appreciated. https://t.co/K7Lh3hIrl5
biorxiv.org
Nanopore sequencing measures ionic current during the translocation of DNA, RNA or protein molecules through a nanoscale protein pore. This raw current signal data can be ‘basecalled’ into sequence...
squigualiser has become more powerful 🚀. This signal pileup view of @nanopore r10 DNA signals will deepen your read analysis on IGV. https://t.co/L0dOzTPOaV
0
6
22
Take a break from worrying about the AllOfUs Umap and check out Squigualiser- our new tool for @nanopore signal data exploration. Great engineering by @hiruna72 and @Hasindu2008 as always 🔥🔥 https://t.co/CpIUEkXDwC
biorxiv.org
Nanopore sequencing measures ionic current during the translocation of DNA, RNA or protein molecules through a nanoscale protein pore. This raw current signal data can be ‘basecalled’ into sequence...
1
10
31
Nice to play a small part in this. Fantastic work by @htanudisastro & @hdashnow 🧬🧬🧬
Our review article on sequencing & characterising short tandem repeats (STRs) is out in @NatureRevGenet! We delve into short read & long read STR genotyping tools and applications in rare disease & population genetics🧬 https://t.co/nHFomYSxLQ
@GenTechGp @hdashnow @dgmacarthur
0
1
10
Bioinformatics behind: this is an example big project where we used S/BLOW5 ecosystem to efficiently, economically (saving compute, time, energy & money), consistently process & reliably archive 141 nanopore signal datasets (~100TB) on Australia's NCI-Gadi @NCInews supercomputer.
Happy & humbled to see our work on structural variation in Indigenous Australians out in @Nature. Deep gratitude to the Aboriginal communities whose leadership & engagement made it possible, & to our @NCIG2013 collabs @hardiprpatel @azure_peacock 🪃🧬🪃 https://t.co/AxKVA53kw6
1
10
33
Great summary! @BlackOchreOmics @BiancaNogrady “We need to build relationships, we need to prove we are careful & considered stewards … & won’t make moves without communities being across the decisions … & have their voice heard in that process,”
nature.com
Nature - In collecting genomic data for Indigenous Australians, scientists hope to expand knowledge of human genetic diversity and improve health for this group.
0
2
10