Yanik Weber
@YanikWeber
Followers
44
Following
73
Media
4
Statuses
23
A biotech enthusiast with a knack for gene editing, exploring the realms of synthetic, chemical biology and directed evolution.
Joined November 2020
RT @talasandris: Our paper describing an RNA-LNP prime editing strategy for treating phenylketonuria was published today in @natBME (1/7).h….
nature.com
Nature Biomedical Engineering - A transient in vivo prime editing strategy is developed for the liver by delivering the prime editor as mRNA encapsulated in LNP.
0
10
0
RT @hollfelderlab: Remkes Scheele et al. make a new reaction accessible to large scale screening by tuning a DNA-aptamer to provide a fluor….
pubs.acs.org
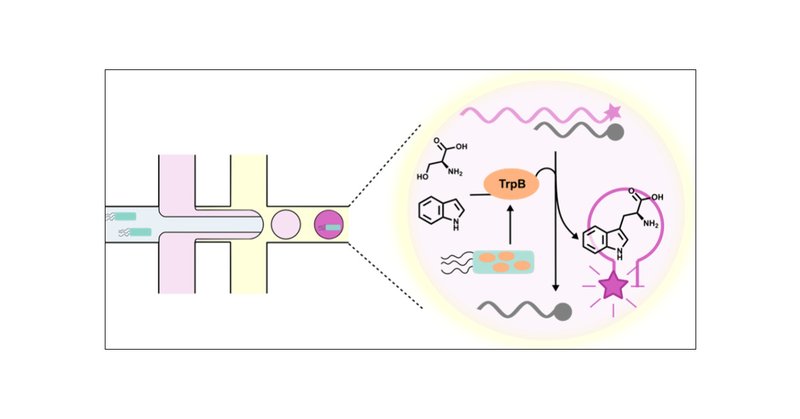
Tryptophan synthase catalyzes the synthesis of a wide array of noncanonical amino acids and is an attractive target for directed evolution. Droplet microfluidics offers an ultrahigh throughput...
0
5
0
RT @schwanklab: Together with the lab of Niek van Til from the UMC Amsterdam we treated vanishing white matter mice by in vivo base editing….
0
8
0
RT @schwanklab: Here we demonstrate that OrthoRep can be employed to evolve genome editors - Congrats to Yanik and many thanks to all colla….
0
22
0
Thanks to all the collaborators @AnastasiaIvascu @BoeckDesiree @tanjarothgangl @nicopmat @marquark @KisslingLucas and others for the support and help on this amazing project and I’m looking forward to next directed evolution endeavors.
0
0
1
It is my pleasure to announce the latest publication from our lab @schwanklab in collaboration with the lab of Martin Jinek @MartinJinek and Norbert Pardi, where we adapted OrthoRep for the directed protein evolution of prime editors.
3
5
37
RT @schwanklab: Big congrats to @tanjarothgangl @BoeckDesiree @lukas_villiger to the Pfizer Forschungspreis, well deserved!.
0
2
0
RT @schwanklab: Here we successfully altered mouse behavior through in vivo prime editing of the beta-1 adrenoceptor in the brain – Big con….
biorxiv.org
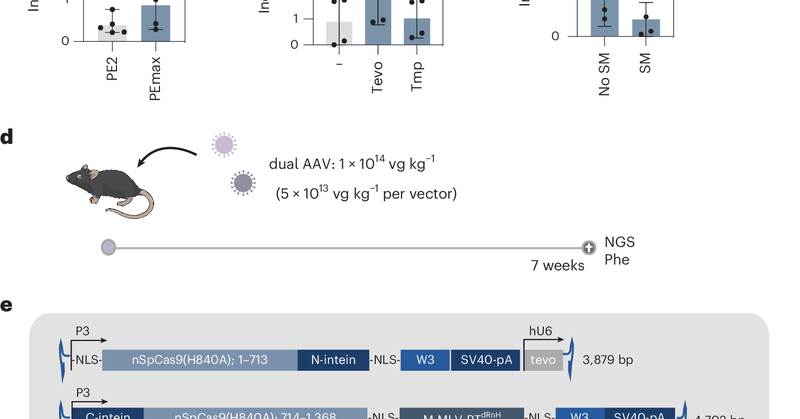
Prime editing is a highly versatile genome editing technology that holds great potential for treating genetic diseases[1][1], [2][2]. While in vivo prime editing has recently been conducted in the...
0
12
0
RT @biorxivpreprint: Click editing enables programmable genome writing using DNA polymerases and HUH endonucleases .
0
11
0
RT @nicopmat: 🚨New preprint🚨Today, we're excited to share our latest study, "Predicting prime editing efficiency across diverse edit types….
biorxiv.org
Prime editing is a powerful genome editing technology, but its efficiency varies depending on the pegRNA design and target locus. Existing computational models for predicting prime editing rates are...
0
26
0
RT @pdhsu: Just shared at @KeystoneSymp a new @ArcInstitute discovery of the bridge RNA recombinase mechanism: a new class of natural RNA-g….
0
327
0
RT @schwanklab: Happy to share our recent advances in developing clinically viable in vivo #CRISPR prime editing approaches, which we just….
biorxiv.org
Prime editing is a versatile genome editing technology that does not rely on DNA double-strand break formation and homology-directed repair (HDR). This makes it a promising tool for correcting...
0
13
0
RT @NobelPrize: BREAKING NEWS.The 2023 #NobelPrize in Physiology or Medicine has been awarded to Katalin Karikó and Drew Weissman for their….
0
26K
0
RT @schwanklab: evoCjCas9 - a new addition to the genome editing toolbox! Big congrats to @LSchmidheini and everyone from the @schwanklab a….
0
8
0
RT @nicopmat: Excited to see our study "Predicting prime editing efficiency and product purity by deep learning" published in @NatureBiotec….
0
27
0
RT @gmdemarchis: Congratulations to Liliane Kriemler on a great talk on apo(b): if LDL is <1.8mmol/l (<70md/dl) but apo(b)>65 mg/dl ❗️, con….
0
3
0