Victor Klein-Sousa
@VKleinSousa
Followers
249
Following
1K
Media
7
Statuses
91
PhD Fellow @cphbiosciphd @NMITaylorLab @UCPH_CPR 🇩🇰 | Structural biology 🤝 phages 🤝 protein folding | from 🇧🇷 | Physicist by training ✏️ (he/him)
Joined January 2021
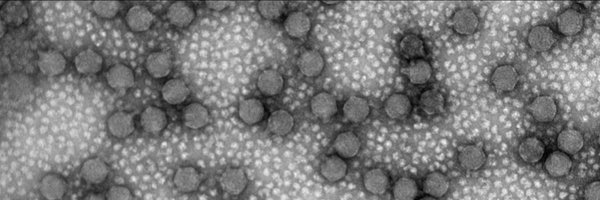
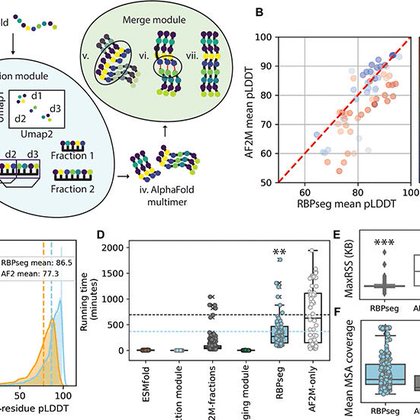
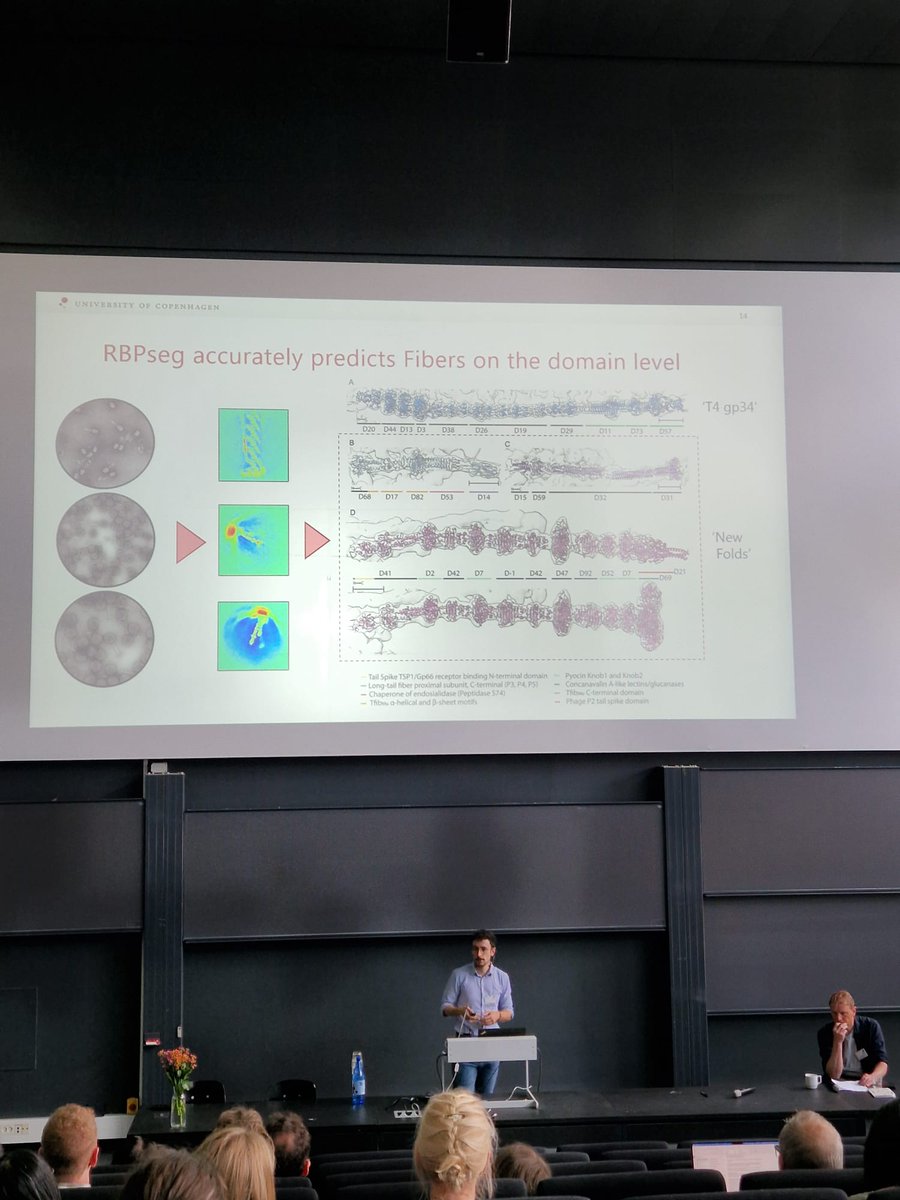
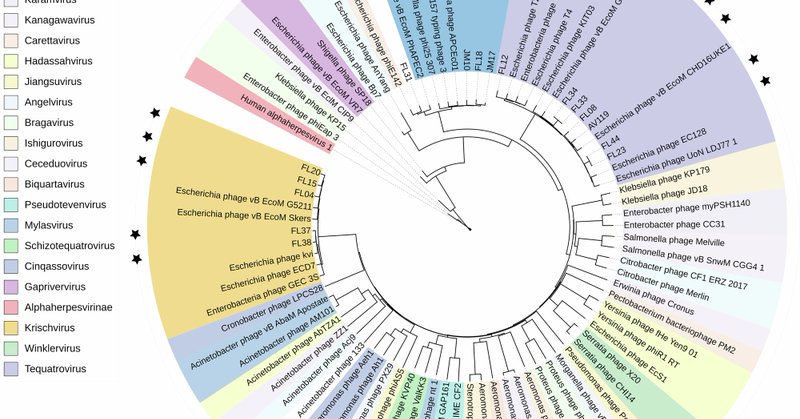
Excited to see our work published in @ScienceAdvances!. We present RBPseg, a method for structural prediction of #phage receptor-binding proteins. We applied it to diverse tail fibers and validated the predicted structures using #cryoEM. Read here:
science.org
RBPseg enables accurate modeling of tail fiber structure, providing the first comprehensive tail fiber structure atlas.
4
23
51
I wonder if prophages would be considered doping in a bacterial Olympics. Fascinating paper.
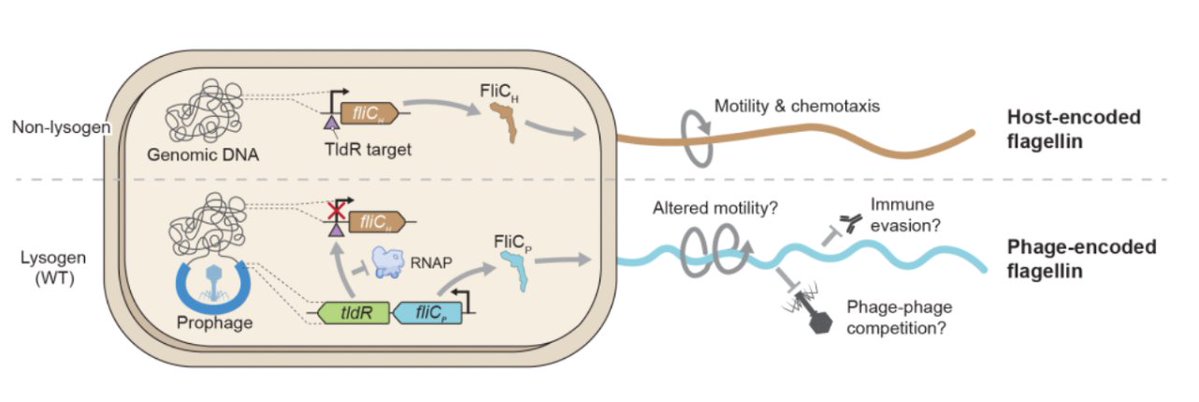
1/16 New pre-print from the Sternberg Lab!.We uncover how temperate phages can use RNA-guided transcription factors to remodel the flagellar composition of their bacterial host and enhance their fitness. Find the preprint and full story here:
0
0
4
RT @RafalMostowy: What determines who a phage can infect?. We tackled this question for temperate phages of Klebsiella — a bacterial pathog….
0
29
0
RT @rishwanth_raghu: Excited to present CryoBoltz ❄️⚡, a multiscale guidance approach for steering AlphaFold3/Boltz-1 to sample structures….
0
52
0
@ScienceAdvances @AritzRoa @CloudyKielkopf @sofos112 @NMITaylorLab @DanishVOM I have just released a new RBPseg version (v.1.1.1) with a few corrections to the merging module and some additional examples on how to run it. Special thanks to @VRajachandran for bringing this issue to my attention. If anyone finds trouble running RBPseg, please reach out!.
0
0
3
RT @NMITaylorLab: Exited to share that our RBPseg paper has just come out in @ScienceAdvances. Huge cograts to first author @VKleinSousa an….
science.org
RBPseg enables accurate modeling of tail fiber structure, providing the first comprehensive tail fiber structure atlas.
0
4
0
RT @Patrick18287926: Our latest work is out: we designed dual GLP1R/GCGR agonists—cyclic peptides that activate both metabolic receptors, e….
0
35
0
@ScienceAdvances @AritzRoa @CloudyKielkopf @sofos112 @NMITaylorLab This came at a great time, as I'll be presenting it at @DanishVOM tomorrow morning. Looking forward to seeing many of you there.
2
1
3
@ScienceAdvances Thanks again to the co-authors @AritzRoa @CloudyKielkopf @sofos112 & @NMITaylorLab.
1
0
1
@ScienceAdvances You can explore the RBPseg code here:. We’ve also added a module to classify new fibers, and a validation model to help assess prediction reliability - going beyond standard AlphaFold metrics.
github.com
RBPseg: A Tool for Tail Fiber Structure Prediction - VKleinSousa/RBPseg
1
0
1
RT @aipulserx: How can artificial intelligence overcome the computational limitations of predicting complete bacteriophage tail fiber struc….
0
2
0
RT @Rooshanie: Contractile injection systems are used by bacteria as tools to survive, compete and interact with their environment, check o….
0
2
0
I’ve been exploring interactive networks to visualize #protein structure contacts — super helpful for large viral complexes. I put everything together in a Google Colab in case it helps others too. Here’s how it looks for the phage T4 baseplate:
1
2
16
I'm happy to see our collaboration with @phagebioUCPH @LNdsted out on npj viruses! I had a fun time modelling T-even #phages adhesin proteins. Verifying that we can use #AF2 bias to select true binders was even cooler. Congrats to Veronika Lutz and all
nature.com
npj Viruses - Gp38 adhesins of Straboviridae phages recognize specific extracellular loops of outer membrane protein receptors
2
1
17
RT @JamesBeanBio: Excited to announce that collaborative work between the Bostina lab (@mihnea_bostina) and PHI lab (@PeterFineran) at @ot….
nature.com
Nature Communications - The cryo-EM structure of phage φTE is presented, revealing a distinct neck topology, tail sheath baseplate organization and oligomeric state of the tape measure...
0
12
0
RT @art_egorov: 💛LoVis4u - a visualisation tool for comparative genomics and sequencing coverage profiles is now published! .See our paper….
0
78
0
RT @Patrick18287926: Two weeks left to apply to the postdoc position with us: Are you passionate about AIxLab inter….
0
7
0
RT @DanishVOM: 🚀 Calling all early-stage researchers! Need support to attend the First Symposium of the Danish Viruses of Microbes Network….
eventsignup.ku.dk
0
4
0