Sternberg Lab
@SternbergLab
Followers
1K
Following
7
Media
49
Statuses
81
News from the Sternberg lab at Columbia Univ., HHMI. Posts are from lab members and not Samuel Sternberg unless signed SHS. Posts represent personal views only.
New York, NY
Joined October 2017
New pre-print(s) from the Sternberg Lab in collaboration with @LeifuChangLab! We uncover an unprecedented molecular mechanism of CRISPR-Cas12f-like proteins, which drive RNA-guided transcription independently of canonical promoter motifs. Full story here:
4
50
195
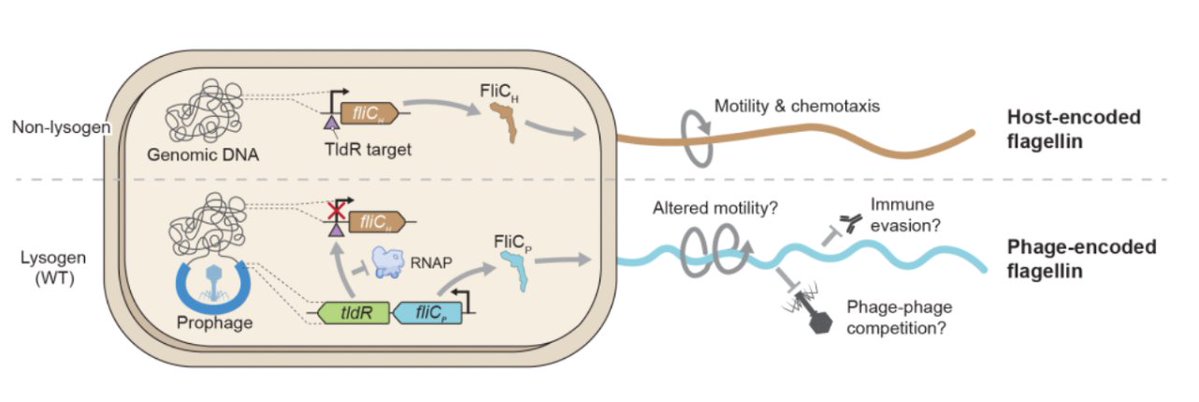
10/16 To try to link these phenotypic observations, we teamed up once again with @IsraelF96135088 and decided to inspect the structures of the prophage flagellin in comparison to their host counterpart.
1
0
3