Tirosh Lab
@TiroshLab
Followers
2K
Following
60
Media
52
Statuses
139
Cancer Systems Biology
Weizmann Institute of Science
Joined July 2019
Congrats Alissa!! Highly recommended for students
I'm thrilled to share that I'll be starting my lab ( https://t.co/h1qdyhRdgN) at the Lunenfeld-Tanenbaum Research Institute @SinaiHealth and joining @MoGen_Grad at the University of Toronto as an assistant professor this fall. 1/
0
0
11
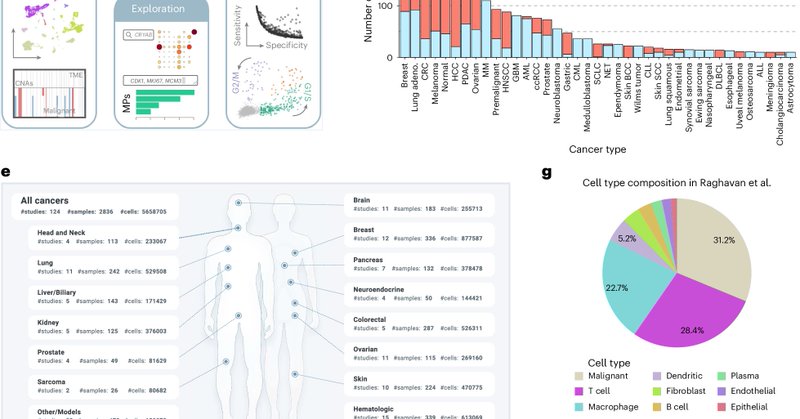
👉The Curated Cancer Atlas. Impressive: “124 scRNA seq datasets for >40 cancer types, with improved data annotations, visualizations and exploration”. From Michael Tyler and @TiroshLab
@WeizmannScience
nature.com
Nature Cancer - Tyler et al. introduce a curated collection of 124 multicancer, single-cell RNA-sequencing datasets with 2,836 samples and present recurrent cancer cell expression metaprograms,...
0
6
18
Important publicly available resource from @TiroshLab @WeizmannScience in @NatureCancer The Curated Cancer Cell Atlas provides a comprehensive characterization of tumors at single-cell resolution https://t.co/9fiBGjFdwU 124 single cell datasets across 40 cancers, >2800 samples.
nature.com
Nature Cancer - Tyler et al. introduce a curated collection of 124 multicancer, single-cell RNA-sequencing datasets with 2,836 samples and present recurrent cancer cell expression metaprograms,...
2
50
186
1/ Truly excited to share our new study that I had to privilege to co-lead during my PhD alongside great friends and collaborators @M_Nomura, Kevin C. Johnson and Luciano Garofano, which was published @NatureGenet! https://t.co/vFNeqB6eqm
nature.com
Nature Genetics - Comparison of paired primary and recurrent glioblastomas at the single-cell transcriptomic level describes molecular and cellular trajectories associated with tumor recurrence,...
3
29
106
1/ Thrilled to share our TWO back-to-back papers published in Nature Genetics today! https://t.co/n9nxmr9dcM ,
nature.com
Nature Genetics - Comparison of paired primary and recurrent glioblastomas at the single-cell transcriptomic level describes molecular and cellular trajectories associated with tumor recurrence,...
15
42
146
Now online in @CD_AACR: #Glioblastoma-Cortical #Organoids Recapitulate Cell State Heterogeneity and Intercellular Transfer - by Vamsi Mangena, @RChanoch, Paola Arlotta, @tiroshlab, @MarioSuva, & colleagues https://t.co/kTHJe4cfE2
@broadinstitute @weizmannscience @MGHCancerCenter
1
20
67
A wonderful collaboration with @MarioSuva lab, @MarianNeidert, co-led by @alissacg, @GaliliNoam and @RouvenHoefflin, with thanks to all authors for their contributions! #spatialbiology #GBM #Visium #CODEX #PhenocyclerFusion #hypoxia 13/end
5
7
40
The layers model is consistent with histology and IVY-GAP annotations but provides further granularity, dissecting the cell state and cell type composition of classical features and exposing the long-distance effects of hypoxia. 12/
1
1
10
We plotted the consensus interactions as a graph and from this a higher-order organization emerged: 5-layers of cell states, consistent with a gradient of hypoxia effects. 11/
1
1
11
Next, we measured associations between pairs of states at multiple levels of resolution. Many spatial associations recurred robustly across samples and scale, which we termed consensus interactions. Other recurring interactions were distance-dependent. 10/
1
2
9
Structure was associated with abundance of hypoxic cancer cells. While hypoxia is known to induce a local structure seen by histology, we find that hypoxia conferred a global organization to cancer cells that far exceeded the scale of structure visible by H&E. 9/
1
1
10
Over larger regions, all states tend to be either structured, with most spots surrounded by spots of similar composition, or disorganized (scattered). Based on histology, disorganization is considered the glioma default, yet we find significant structure in many samples. 8/
1
1
5
At the spot level, states tend to cluster most with themselves, such that spots are dominated by 1 or 2 cell states. 7/
1
1
7
Visium covers most genes but lacks true single-cell resolution. Therefore, we designed an Ab panel based on sc signatures & used CODEX as a validation. We could also generate Visium-sized pseudospots over the CODEX data to ask questions about their ground truth composition. 6/
1
2
19
Spatially-derived MPs included those we previously identified by scRNA-seq as well as new MPs unique to the spatial data. Several of the new MPs were spatially associated with hypoxia. 5/
2
2
7
We derive the composition of each sample directly from the spatial data by identifying recurrent groups of genes that are coherently expressed – metaprograms (MPs) – that vary within each sample and represent a single cell state or cell type. 4/
1
2
11
To answer this question, we profiled glioma samples by spatial transcriptomics & proteomics. For analysis, we integrated our Visium dataset with samples from Ravi et al. ( https://t.co/xw0C8jmT9y). 3/
2
2
15
Previously, we defined the core cancer cellular states for GBM by scRNA-Seq. How do the cell states relate to one another in space? Are spatial associations between states stochastic or do recurrent patterns emerge? 2/
1
2
7
Excited to share our new paper in @CellCellPress on spatial organization in glioblastoma https://t.co/XHcI7WQyot. We combine spatial transcriptomics (Visium) and proteomics (CODEX) to reveal five-layer organization of cellular states and a role of hypoxia as tissue organizer. 1/
19
158
657