Microbiome Research
@SunagawaLab
Followers
2K
Following
107
Media
11
Statuses
85
Microbiome Research Lab at the Institute of @Microbio_ETH Zurich | Tweets by lab members
Zürich, Switzerland
Joined March 2019
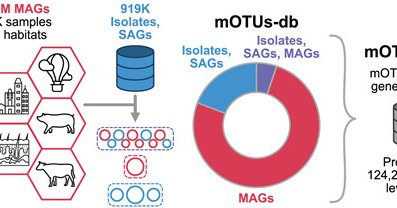
The mOTUs4 genome database: 2.83Mio systematically reconstructed MAGs and >900K isolate and single cell-assembled genomes, all clustered into 124K species-level clusters. Paper out at @NAR_Open: https://t.co/tviEEeLObk 1/5
academic.oup.com
Abstract. Determining the taxonomic composition (taxonomic profiling) is a fundamental task in studying environmental and host-associated microbial communi
1
15
30
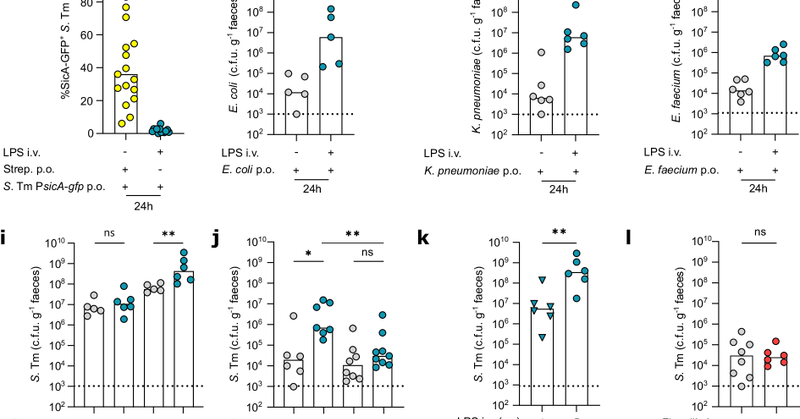
Great to see this study out that members of our lab contributed to: systemic immune activation (through lipopolysaccharides) promotes 100-10,000 fold expansion of opportunistic pathogens in the mouse gut -
nature.com
Nature Communications - Acute systemic immune activation may affect gut-luminal colonisation dynamics. Here, the authors show that LPS exposure in mice elevates gut-luminal oxygen species levels,...
0
0
1
Check out this interview with @PietroTedesco on the work that he is contributing to #BlueRemediomics - an EU horizon Project that we are also a part of!
🎙️🔬We spoke with #BlueRemediomics researcher @PietroTedesco from @SznDohrn who focuses on identifying #compounds & #enzymes with high potential for pharma or cosmeceutical applications. 📹 Watch the video with Pietro below! 👉 Read the interview here: https://t.co/le5sEA9TMR
0
0
1
Incredibly happy and grateful to see this major part of my PhD thesis published. Thanks to all collaborators! @pfeilmes @aasintsova @SunagawaLab @Vorholt1
1
1
3
Last week we hosted a group of schoolchildren in our lab to discover environmental microbes during the National Future Day @Zukunftstag at @ETH_en. We had a lot of fun isolating bacteria and fungi and studying them under the microscope
0
1
5
Letzte Woche war eine Gruppe von Schulkindern in unserem Labor auf Spurensuche nach Umweltmikroben während des Nationalen @Zukunftstag an der @ETH_en. Wir hatten viel Spass beim Isolieren von Bakterien und Pilzen und beim Programmieren von ihrem eigenen Bild.
0
0
0
Congratulations to @marija_dmit, @hjruscheweyh, @Samuel_BIO2810, @aasintsova, @pangenomics, and @ZellerGroup 🎉 5/5
0
1
2
Users can interactively search, query, and download sequence data from genomes and mOTUs on the database website: https://t.co/tckM2AYAaj 4/5
1
0
1
The species-level taxonomic units (mOTUs) have been annotated using GTDB taxonomy and are available for taxonomic profiling using the mOTUs4 tool: https://t.co/1EOt0sLI3S 3/5
1
0
1
MAGs were reconstructed 118K metagenomic samples from over 70 different environments using a standardized workflow, which includes multi-sample coverage information for binning scaffolds. 2/5
1
0
1
This work is here thanks to @emily_fogarty11, whose dedication turned a surprising observation into a piece of science. But many others helped us push it forward. @sandralmclellan, @KarenLolans, @mschecht_bio, @AmyDWillis, @SunagawaLab, Laurie Comstock et al, THANK YOU ALL :)
1
2
13
A reminder for ECRs who are looking for an opportunity to form their own group to study contemporary questions in marine sciences in Germany with 5-years of support: https://t.co/uIJfJ8CdV0
0
38
62
New computational tools from @SunagawaLab @NCCRMicrobiomes 👏 mBARq: a versatile and user-friendly framework for the analysis of DNA barcodes from transposon insertion libraries, knockout mutants and isogenic strain populations
biorxiv.org
DNA barcoding has become a powerful tool for assessing the fitness of strains in a variety of studies, including random transposon mutagenesis screens, attenuation of site-directed mutants, and...
1
7
17
I'm excited to share that I've accepted a position as Professor for #environmental #microbiology at @uniinnsbruck in Austria. My lab will focus on microbial omics of uncharacterised taxa and on plastic biorecycling. Will post job ads shortly - for 2x PhD and 1x postdoc
36
58
643
Predicting interactions in plant microbiomes made possible. Metabolic modelling sheds light on bacterial interactions in planta, see the recent work by Vorholt Lab @ETH_en and @realLCSB at EPFL. More: https://t.co/hzWV3ogfjp
0
11
21
What is #EMBLtrec about? Why did we embark on a journey along the European coastlines? What are the scientific questions we are studying? Watch our new video to find out! https://t.co/F1jwRHPkd4
@embl @TaraOcean_ @EMBRC_EU 📸 Kinga Lubowiecka / EMBL
0
13
39
We're hiring for a postdoc position focused on metabolic modeling. Please share widely.
4
181
228
The huge success of #HIFMBsymposium's session 3 on the multi-data matrix behind functional biodiversity are attributed to: Linda Amaral Zettler @plasticphere, Shinichi @SunagawaLab, Kirsten Küsel @iDiv and Boris Koch @AWI_Media #thankyou #microorganisms #chemodiversity
0
2
6
So many creative ideas and forward looking plans at the 2nd symposium on federating bioinformatics tools and resources last week. Truly thrilled to be part of this group of scientists
0
2
5
📣Open postdoc position📣 in our group @Microbio_ETH to work on marine protist-microbe interactions. Lots of opportunities for networking! See job ad for details: https://t.co/5RI5R6RbzL
0
10
22