the real LCSB
@realLCSB
Followers
335
Following
143
Media
36
Statuses
223
Laboratory of Computational Systems Biotechnology led by @Vassily_13 Databases: https://t.co/kdLhSi2CbQ (NEW: ReKinDLE) Opensource: https://t.co/RJeBF8fVn4
Lausanne, Switzerland
Joined August 2017
This work by Toumpe, Masid, Hatzimanikatis & Miskovic (@epfl_en LCSB) provides a resource for exploring metabolic regulation, vulnerabilities, and precision oncology. All models are openly available. #systemsbiology #cancermetabolism #multiomics #precisiononcology #openscience
0
1
0
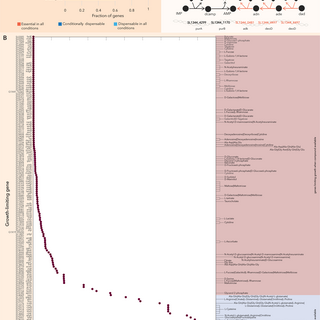
By integrating multi-omics data with enzyme kinetics, we generated a population of near–genome-scale kinetic models that capture BRCA1-mutant vs wild-type ovarian cancer physiology. These models reveal network-wide control principles & drug-response dynamics.
1
1
0
To understand how cancer cells dynamically reprogram metabolism, we built the largest dynamic reconstructions of human & cancer metabolism to date — >2,300 reactions across major pathways. Comparable in scale to genome-scale E. coli models!
1
0
0
🚀 New preprint from @epfl_en’s Laboratory of Computational Systems Biotechnology (LCSB)! We present “Multi-omics–driven kinetic modeling reveals metabolic vulnerabilities and differential drug-response dynamics in ovarian cancer.” 🔗 https://t.co/EaqYTn8bRq
1
1
0
Our review made the cover in ACS Synthetic Biology! 🎉🧬 We explore how large-scale kinetic models are shaping the future of metabolic engineering — check it out here 👉 https://t.co/GsO6bv8JTF Big thanks to the team and @ACSPublications! #MyACSCover #SyntheticBiology
pubs.acs.org
Recent advances in mechanistic kinetic modeling now enable detailed descriptions of metabolism across the universe of living organisms. We highlight the potential of these developments to drive...
0
2
3
Excited to share our new preprint, where we present NIS, a framework that distills GEMs into interpretable modules, enabling direct cross-species comparisons of fueling pathways, biosynthesis, and environmental exchanges. Congratulations to the authors!
👏 A big thank you to my co-authors Meric Ataman & Vassily Hatzimanikatis for this great collaboration! 🔍 Read more:
0
1
2
Our latest work on Salmonella metabolism in the murine gut is now at @PLOSCompBiol ! Congratulations to the authors @EVayena, @FuchsiLea, @HomaMP4, Konrad Lagoda, Bidong Nguyen, Wolf D. Hardt, and @vassily_13! https://t.co/KmXzKr7IQR
journals.plos.org
Author summary Nontyphoidal Salmonella strains are among the most ubiquitous enteropathogens that contribute significantly to global morbidity and mortality, posing a substantial burden on public...
0
1
2
This work showcases one of the few successful integrations of kinetic modeling and experiments to optimize cell factories. It highlights the power of NOMAD ( https://t.co/2zbJiEMFWi) in accelerating and improving strain design!
nature.com
Nature Communications - No consensus exists on the computationally tractable use of dynamic models for strain design. To tackle this, the authors report a framework,...
0
3
1
Excited to share our preprint on boosting p-coumaric acid production in S. cerevisiae! 🎉 In collaboration with Irina Borodina's group (@Irina__Borodina), we used our NOMAD framework for strain design strategies. Read more:
biorxiv.org
The use of kinetic models of metabolism in design-build-learn-test cycles is limited despite their potential to guide and accelerate the optimization of cell factories. This is primarily due to...
1
2
1
A new baby from our creative workshop:
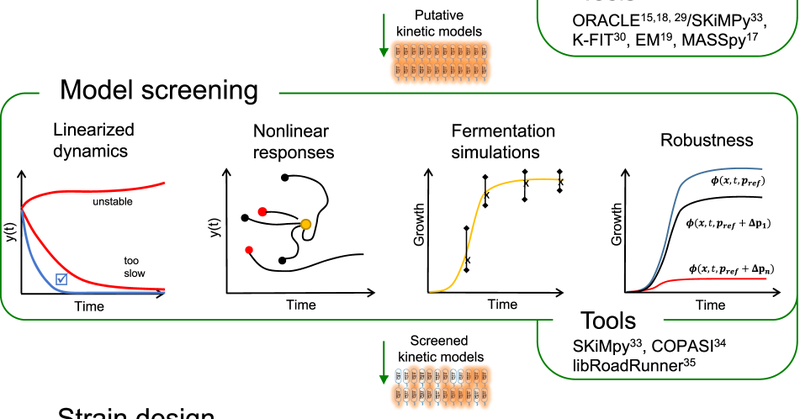
Want to efficiently create large-scale dynamic models of metabolism that fit experimental data and reliably predict metabolic responses to various perturbations? Our new method does just that! Check it out here: https://t.co/d757bhiFC8.
0
1
2
We are thrilled to announce that our generative ML method for kinetic modeling ( https://t.co/VTbFKoArEq) has been accepted to @NatureCatalysis. Congratulations to Subham (@astro_dank), Bharath (@bharathnarayana), Michael (@moret1788), Vassily (@Vassily_13), and Misko (@Misko_L34)
biorxiv.org
Generating large omics datasets has become routine practice to gain insights into cellular processes, yet deciphering such massive datasets and determining intracellular metabolic states remains...
0
4
7
Our latest work is now out at Metabolic Engineering! Congratulations to the authors @oftadehomid and @Vassily_13!
Our new paper describes how computational models can capture the #plasmid metabolic burden and help optimize #recombinant expression of proteins https://t.co/WdQLijNNbe
0
2
6
Congratulations to Bharath (@bharathnarayana), Daniel (@realDRWeilandt), Maria, Misko (@Misko_L34), and @Vassily_13 for this nice work ( https://t.co/7bQJrrm1oH)
0
0
1
Our recent publication in @NatureComms, “Rational strain design with minimal phenotype perturbation” by Narayanan et al., has been showcased in the recent Editors’ Highlights for Biotechnology and Methods focus ( https://t.co/YLmSKMX85k).
1
3
5
Check out our preprint on now on bioaRxiv!!! Congratulations to all the authors! @oftadehomid @aslisahin2205 @EVayena @vassily_13
A preprint of our recent work is now available! In this paper we present a framework to infer nutrient competition and cross-feeding in microbial communities. Many thanks for the support we received from @NCCRMicrobiomes. https://t.co/D1GYdTWMSd
0
2
4
Our latest work on metabolic engineering using kinetic models and process systems engineering methods is out https://t.co/7bQJrrm1oH! Congrats to Bharath (@bharathnarayana), Daniel (@realDRWeilandt ), Maria, Misko (@Misko_L34), and @Vassily_13 .
0
10
23
Our latest work on metabolic engineering using kinetic models and process systems engineering methods is out https://t.co/7bQJrrm1oH! Congrats to Bharath (@bharathnarayana), Daniel (@realDRWeilandt ), Maria, Misko (@Misko_L34), and @Vassily_13 .
0
10
23
Congratulations to all the authors for this fantastic achievement! Homa (@HomaMP4), Anastasia (@AnastasiaSves), Misko (@Misko_L34) and Vassily (@vassily_13) !!
0
0
1
Curious and want to dive deeper? Check out our preprint on Bioarxiv:
biorxiv.org
Novel sequencing techniques and biochemical pathway prediction resources provide a wealth of data on novel proteins and computationally predicted enzymatic reactions. Accurate matching of protein...
1
1
1