Ragothaman Yennamalli
@StructBioinfo
Followers
2K
Following
50K
Media
1K
Statuses
12K
Sr. Asst Prof @SastraUniv | Bioinformatician, He/Him. co-organizer #YIM2024, Asso.Ed @csb_journal, Spl Issue Ed: @BiophysJ Ed: BMC Bioinfo, Prot Sci, Sci Rep
India
Joined May 2013
🚨🚨🚨Our latest work @SastraUniv got featured in @NatureInd!!!🚨🚨🚨 https://t.co/YgsTO6GtE2
@s_shricharan @SankarMaheshR @SubachandranJK The work was funded by @ugc_india
nature.com
Nature India - Knowing how specific enzymes enable heat tolerance could cut industry costs
2
5
64
My woes continue for another manuscript. This is more than a month after submission. My hunches for the string of bad luck are: 1. It's me! 2. Summer just got over and everyone is under a hangover and not ready to do their "work". 3. Current Solar activity 4. Some paranormal act!
After a month, I got this message from the editor and the manuscript is back to me. I did suggest referees, at submission. Seriously, what are my options now? Write back with a few more suggestions? Openly seeking advice. 🙏🏽 TIA!
4
0
6
What's the go to CRISPR/Cas9 kit for practical/demonstration purposes?
1
0
3
Sad to hear that David Baltimore passed away! ☹️His book Molecular Cell Biology authored with Lodish was one among my favorites, after Genes and Molecular Biology of the Cell. RIP!
Legend David Baltimore died yesterday. He understood the way things should work: "the real contribution of @MIT is that it doesn't take itself too seriously. It takes ideas seriously, but the people are relatively informal. They are not self-aggrandizing the way academics can be.
0
0
9
Himalayan museum before and after renovation. Still some work needs to be completed!
6
8
35
Do you know what this is? At first glance it’s just an old wooden board. But it’s hiding a classroom story from ancient Egypt… one that feels surprisingly familiar even today. Read on 1/10
2
19
88
Congratulations to Dr.Shankar Sriram for receiving National Teachers Award from Hon'ble President of India in the presence of @dpradhanbjp ji. This @EduMinOfIndia Award is recognition of your exceceptional teaching capabilities that have +vely impacted 1000s of students' careers
2
14
113
Thank you Prof Sood and Dr Vinod Paul for Launching the Book At War with the Single strand edited by me Prof Padmanaban and Dr NK Arora .20 chapters contributed by key players document the #COVID journey.Details of the book are at https://t.co/YgNxFlFjDb
@NITIAayog
Prof. Ajay Kumar Sood, @PrinSciAdvGoI and Dr. Vinod Paul, Member (Health), @NITIAayog, launched the book titled “At War with the Single Strand - How Indian Science Fought the COVID-19 Pandemic” on 4th September 2025 at the India International Centre, New Delhi.
5
14
47
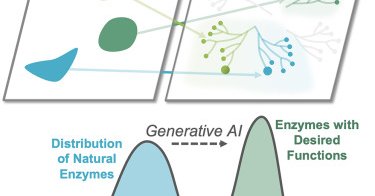
In this perspective, we discuss a vision for AI-driven #enzyme discovery to unveil a world of enzymes that transcends biological evolution and perhaps offers a route to genetically encoding almost any chemistry. https://t.co/rfGwDHHJdC
cell.com
Artificial intelligence is revolutionizing how we engineer enzymes, enabling the creation of biocatalysts beyond those shaped by evolution. In this perspective, we discuss a vision for how artificial...
1
30
146
🧵Regex (regular expression) can save your bioinformatics analysis. Most wet lab people are not aware of it But it’s the difference between chaos and clarity. Here’s how.
3
11
86
After a long time, good news is coming soon...Watch out for this space...
1
0
17
Traveling sans toddler so devouring this book! Thanks @jenheemstra for sharing your wisdom #AcademicExcellence #AcademicChatter #AcademicTwitter
1
4
19
As I complete 1 year at IISER Bhopal, I take some time to reflect upon some important aspects of a faculty career (especially in the Indian academia context). Since my entire training (PhD and postdoc) was in the US, I faced some initial "culture shock" but soon I adapted. (1/14)
9
6
181
Happy to share that I am now part of the Editorial Board of Scientific Reports @SpringerNature
15
1
133
What everyone thinks happens in my lab! What is actually happening in the lab!
0
0
5
I think you can be a good person and a good scientist. I will die on this hill.
11
49
426
A good friend Ravi @storyrulesindia has this fascinating new book I will recommend checking out. If you find it interesting might also be worth bringing this to your class. https://t.co/z5e0Txtllg
Hallelujah! ‘Story Rules’, my first book is available for pre-orders! Story Rules is written with the aim of being the most comprehensive how-to manual on the craft of storytelling. It provides 31 timeless storytelling techniques to help you: - Be clear: Sharpen your messages,
0
2
5
*Save the Date for Young Investigators' Meeting 2026* IndiaBioscience is excited to announce that our next YIM will be held from 2-6 March 2026 in Pune, Maharashtra. We invite all the Young Investigators working in Life Sciences and Biotech in India to block their calendars and
3
21
47