Shai Carmi
@ShaiCarmi
Followers
4K
Following
4K
Media
677
Statuses
5K
Find me at @shaicarmi.bsky.social
Jerusalem, Israel
Joined February 2019
An ad campaign in the NYC subway that urge people to select the traits of IVF embryos is proving to be a test of anti-discrimination laws.... which apply to ads on public property. Fascinating as a breakthrough example of direct-to-consumer ""eugenics"" Story link👇
1
7
23
A look at the process, history, and ethics of a potentially revolutionary new technology. By @Rosalind_Arden_. https://t.co/SnaHyJkQdL
quillette.com
A look at the process, history, and ethics of a potentially revolutionary new technology.
3
9
28
Polygenic embryo screening is being marketed commercially – but how do IVF clinicians view it? • General approval is low (12%) For specific uses: • 59% approved of health-related embryo selection • 6% approved of trait-based selection 🧵 Survey findings in NPJ Genomic Medicine
4
9
26
Be part of this exciting campus! Share your data on reproductive aging by submitting your abstract: 🗓️Deadline 16 November 2025 @ESHREsigRG
🧬 Understanding Reproductive Aging: A Critical Conversation! Explore genetic insights & emerging therapies with leading experts 📈9.5 CME credits 📝 Abstract submissions open until 16 Nov 📍Frankfurt, Germany | 🗓️ 4-6 December 🔗 https://t.co/6JqYiT6wtQ
#ESHRE #CampusCourse
0
2
2
New Preprint! Led by @ShaiCarmi and his student Liraz Klausner, we have developed an online risk calculator to explore parameters for polygenic embryo screening (PES). We demonstrate (for the first time), the degree to which real-world live birth rates attenuate PES effects.
3
20
53
One out of many excellent scientific activities @ESHRE is organising for its members and all those interested in #reproductive science. 🧬Discussing the limits in reproductive #genetictesting on 30/9, a webinar organised by @ESHREsigRG
🧩Join #ESHRE SIGs to explore key challenges in #ReproductiveMmedicine! 🔹16 Sep: Implantation & Early Pregnancy: facing barriers 🔹23 Sep: Embryology: fate of “unfit” embryos 🔹30 Sep: Reproductive Genetics: testing limits 🕔17:00–18:00 CEST https://t.co/lUPTKtoPCn
0
3
6
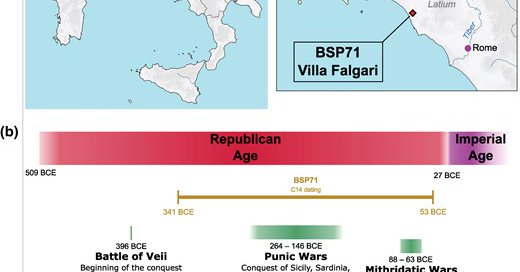
🚨 New paper out in Genome Biology and Evolution! We present new ancient DNA evidence from Central Italy suggesting that Eastern Mediterranean genetic influence predates the Roman Empire. @Benjopop_1977 🧵👇 🔗 https://t.co/2ISI9rYmbc 1/
academic.oup.com
Abstract. Italian genetic history was profoundly shaped by the Romans. While the Iron Age Central Italian gene pool was comparable to that of other coeval
4
14
59
We just published this paper in Genetics in Medicine examining how different populations view and use the concept of "eugenics" when discussing polygenic embryo screening. EUGENICS AND POLYGENIC EMBRYO SCREENING...
1
3
12
6 years ago, we predicted what Denisovans probably looked like. Since then, every new Denisovan fossil - Harbin, Penghu, Xiahe - nailed it💀. ☑️ Harbin: 16/18 features match ☑️ Penghu: 5/6 ☑️ Xiahe: 4/4 See 🧵 https://t.co/bJxa8GbwN1
2
12
62
Last year, we posted a preprint providing the first genetic evidence that the Harbin skull likely belonged to a Denisovan: https://t.co/DSsB6tkkM7 Today, DNA evidence provided the final proof: https://t.co/guMUY7k8lx
3
20
67
Our lab at the Weizmann Institute was hit tonight by Iran We will rebuild and return 💪
487
637
8K
Preliminary program now available for ProbGen 2026: https://t.co/hMWTPXMDLA. We have an amazing set of speakers let by our keynotes Sally Otto and @jkpritch. 1/8
2
21
49
In the US, embryo editing is barely studied, making a CRISPR baby is prohibited, and there's no startups on the scene. Yet...gene-edit tech continues to advance. Now, billionaire Brian Armstrong says he's ready to fund the defining U.S. embryo-editing company. 👇
7
27
119
Highly recommended and wide-ranging read about the reality of Harvard. "The appropriate treatment (as with other imperfect institutions) is to diagnose which parts need which remedies, not to cut its carotid and watch it bleed out."
8
3
37
New publication! @BiologicalPsyc1 We expand on our prior work demonstrating that pleiotropic effects of cognitive GWAS can provide biological insights into schizophrenia GWAS results. We differentiated "concordant" from "discordant" subsets of schizophrenia risk SNPs (1/3)
2
12
27
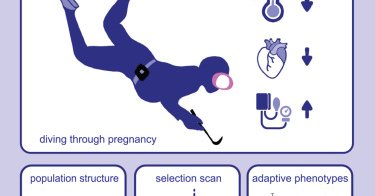
Our paper is finally out: Genetic and training adaptations in the Haenyeo divers of Jeju, Korea: Cell Reports @ras_nielsen @andrewhvaughn
cell.com
Aguilar-Gómez et al. find evidence of evolutionary adaptation with potential relevance to breath-hold diving in the Haenyeo divers of Korea. Genetic variants under selection in this population may...
2
19
43
Latest in #GENETICS, @YileiHuang317, @ShaiCarmi, and @HaraldRingbauer developed a new tool, TTNE, that uses identity-by-descent tracts in large ancient DNA time-series for inferring effective population size trajectories. Read more: https://t.co/cmNnqwzgJr
0
7
12
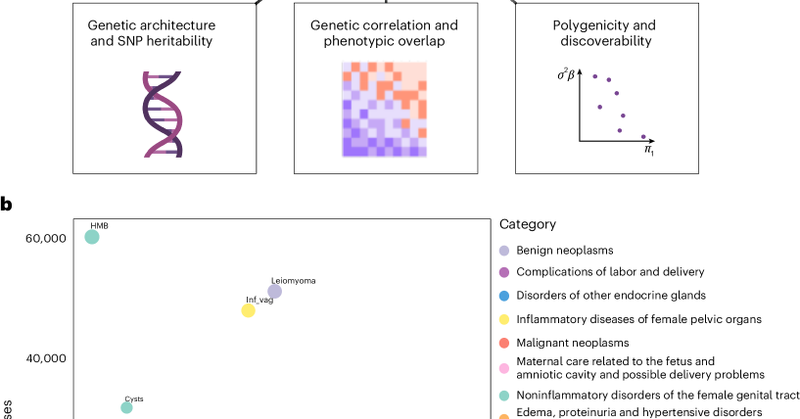
📢 Excited to share our study in @NatureMedicine 👉 https://t.co/eShLKe8xVZ We mapped & characterized genetic risk factors for 42 female reproductive health diagnoses. What we found & why it matters? 👇🧵
nature.com
Nature Medicine - This study provides a cross-trait atlas of genetic and phenotypic associations across 42 female reproductive health diagnoses.
1
5
28
We’re HIRING a clinical ethicist to join the Department of Bioethics & Decision Sciences in the Geisinger College of Health Sciences! There is potential for advancement into a leadership role in the future for a candidate w/preferred qualifications. Pls share widely! (Link next)
1
3
6