Geir Kjetil Sandve
@SandveGeir
Followers
795
Following
117
Media
9
Statuses
227
Professor in bioinformatics/machine learning at University of Oslo, Norway. Follow me at @[email protected] or at @[email protected]
Joined July 2017
Please RT and forward this information to your colleagues, labs, and collaborators who would be interested!
0
0
1
This is a call for experts in #AI, #MachineLearning, #DeepLearning, #AdaptiveImmunity, #Immunoinformatics, #Bioinformatics and #ComputationalBiology to join the challenge and help advance on a crucial #immunology problem with applications in diagnostics and therapeutics.
1
0
1
📢 Announcing the Adaptive Immune Profiling Challenge 2025! Can you predict immune state labels from adaptive immune receptor repertoires? 💰 $10,000 prize pool! 📄 Top 10 teams co-author a @NatureMethods paper! 🗓️ Launches Nov 5 on @Kaggle More Info: https://t.co/WQb8k2RZJT
1
1
2
Another way to beware of intuition around statistics in life science - with hundreds of significant findings, it may seem obvious that at least some must be true. But not necessarily. A thread by Chakri based on a great collaboration with Mamica, Emilie, Manuela and Jessica!
1/12 🧵 Quick thread: a heads-up for anyone working with high-dimensional omics data and multiple testing — FDR correction can sometimes lie to you. A thread based on our recent publication in @GenomeBiology :
0
0
1
This morning, Minister of State @YvanButera launched the Kigali Hub for Spatial & Spatio-Temporal Modelling in Africa — a new go-to center for climate-health modelling, smarter policy, and regional teamwork. The launch wrapped up the 2nd Conference on Spatial Modelling in Africa.
1
54
168
And thanks also to @milenapavl and @KanduriC for their contributions to the work - their critical and constructive input from the early start and throughout the project was crucial. Such curious and supportive attitude is what makes a work environment great!
0
0
4
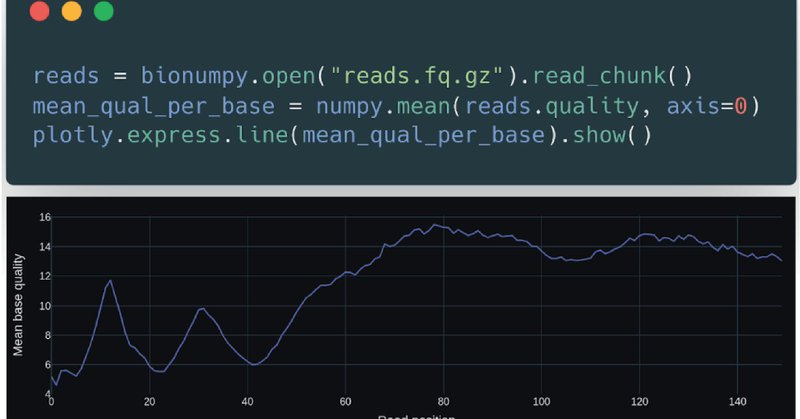
I truly hope the biological community will jump on this fantastic opportunity to save development effort and arrive at computationally efficient (fast and energy saving) code. And thanks to Nature Methods for appreciating generic contributions to science: https://t.co/06MjAmBINx
nature.com
Nature Methods - BioNumPy: array programming for biology
0
0
8
Took a good three years to develop. But had before that hoped for a decade that somebody else would do it.. My deepest gratitude to @knutdrand and @IvarGrytten who did a remarkable effort in landing this complex task in way that comes across very clean and natural for users.2/3
1
0
7
Finally biologists can also use numpy (array programming). Handling e.g. DNA and protein sequences with convenience and speed, like physicists and machine learners for decades have worked with numerical data: https://t.co/06MjAmBINx (1/3)
nature.com
Nature Methods - BioNumPy: array programming for biology
2
42
115
NEWS: @unioslo's Titan magazine published an in-depth article about DHIS2's @wellcometrust-funded Climate & Health work (in Norwegian), featuring colleagues Geir Kjetil Sandve, Zeferino Saugene & Patrick Omiel Okecho. Read more: https://t.co/nX70sw5Q5x
@Saudigitus @HISP_Uganda
0
3
6
Our biophysical cartography of native and human-engineered antibody landscapes is out: https://t.co/L13Z7mVZDy. Study led by @HabibBashour @EvaSmorodina @Jahn_Zhong @pariset_matteo. Great collab with @adaptyvbio @naturalantibody @SandveGeir JTA Andersen.
(1/10) Our work on antibody developability is out! We present a cartography of the developability landscapes of the native and human-engineered antibodies. This study was a team effort of @HabibBashour @EvaSmorodina @pariset_matteo , and @Jahn_Zhong
https://t.co/kIMBjziQAi
2
19
49
One more baby is out! A perspective led by @MaiSpaceHa bring a salt of linguistic in the soup of biological sequence ML models! She describes how antibody sequences of different specificities may form a language, which has implications on the design of future LLM models.
📢 @maispaceha, @PRobertImmodels, @BartlomiejSwia4, @SandveGeir, @victorgreiff et al. explore the potential of linguistic formalization of antibody language to improve rule mining in deep LMs for antibody sequence analysis. https://t.co/BpYjaScgCL ➡️ https://t.co/OvL7J5yZ9f
1
4
10
Very happy to announce that our perspective is finally out in Nature Comp. Sci! Many thanks to Mat Akbar, @PRobertImmodels, @BartlomiejSwia4, @SandveGeir, Dag Haug, and @victorgreiff for the fantastic work.
📢 @maispaceha, @PRobertImmodels, @BartlomiejSwia4, @SandveGeir, @victorgreiff et al. explore the potential of linguistic formalization of antibody language to improve rule mining in deep LMs for antibody sequence analysis. https://t.co/BpYjaScgCL ➡️ https://t.co/OvL7J5yZ9f
1
2
7
Tissue proteomics with clinical value in #celiac disease: - https://t.co/rgqD3HyRFO Great work Anette Johansen and thank you collaborators in particular @SandveGeir for expert ML knowledge! @CeliacDotOrg @UniOslo_MED @Coeliac_UK @AGA_Gastro @coliakiforening
0
2
3
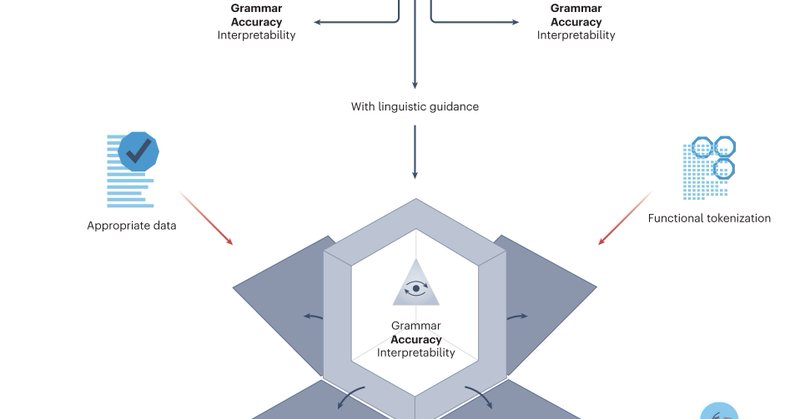
It took us some time to chew on this, and a bunch of (intriguing) nuances came up as we dug deeper, but I think it landed as a quite accessible and useful overview of concerns and opportunities connected to causality and study design for biomarkers! (with AIRR as case)
Our perspective on the generalization of machine learning-identified biomarkers is now published in @NatMachIntell 🎉
0
5
6
It's not often a new method ends up being both faster and more accurate than existing methods, so I believe KAGE2 by @IvarGrtytten and @knutrand (powered by @BioNumPy) will be a great addition to the toolboxes of structural variant discovery researchers https://t.co/mYmDKFjKJK
biorxiv.org
Structural variation is known to play an important and often overlooked role in regulating disease and traits, but accurately detecting structural variants from sequencing data has traditionally been...
KAGE2 is out! Enables very fast and accurate genotyping of structural variants using pangenomes: https://t.co/PHJi7K3fJp. I’ve spent the last 6+ months going deep into the SV rabbit hole, and had some surprises I thought it’s worth to also share (1/6)
0
2
13
(1/8)🎉A fresh preprint in which we present LIgO — a powerful tool to simulate adaptive immune receptor (AIR) and repertoire (AIRR) data for the development and benchmarking of AIRR-based ML 🧵⬇️ https://t.co/0iif0WbUP2
3
21
55
Do you want to model gene regulatory or co-expression networks based on large-scale heterogeneous data, such as data derived from different tissues or cancer types? Have a look at our new normalization method SNAIL 🐌, now out in Bioinformatics 👇: https://t.co/jzJo6NOSpm
2
19
76
1/8 New preprint: generative modeling of AIRR repertoires, the last piece of my PhD, >2 years of work, a project that is very dear to me https://t.co/gwZt4WBuLY
2
9
44
Our perspective on adapting LMs for protein sequences with the help of linguistic knowledge is now out in @NatMachIntell. Led by @MaiSpaceHa, @pandaisikit and @PRobertImmodels. Great collboration with @BartlomiejSwia4, @SandveGeir and Dag Haug.
nature.com
Nature Machine Intelligence - Language models trained on proteins can help to predict functions from sequences but provide little insight into the underlying mechanisms. Vu and colleagues explain...
Happy to share my first preprint in the Immunolingo project “Advancing protein language models with linguistics: a roadmap for improved interpretability”, a perspective on adapting LMs for protein sequences with the help of linguistic knowledge. https://t.co/5U1nzoYdR5
2
17
57