Andrei Slabodkin
@rlyhighvariance
Followers
394
Following
247
Media
9
Statuses
100
ML Scientist at @jura_bio
Oslo, Norway
Joined November 2017
Who would like to synthesize the whole Observed Antibody Space at a cost of several MacBooks?
I’m thrilled to announce: at @jura_bio we’ve constructed a new kind of generative protein model, which allows us to manufacture its generated designs in the real world at petascale. Paper: https://t.co/qgxfNPP30K Blog:
2
7
17
We have trained ESM3 and we're excited to introduce EvolutionaryScale. ESM3 is a generative language model for programming biology. In experiments, we found ESM3 can simulate 500M years of evolution to generate new fluorescent proteins. Read more: https://t.co/iAC3lkj0iV
140
812
3K
(1/7) Curious about what a blood sample can reveal about humoral immunity? Our latest work combined sequencing methods and mass spectrometry to explore BCR and antibody repertoires and assess their synergistic potential. https://t.co/kGQEgTm93q
1
17
61
The IMMREP23 @kaggle #TCR-#epitope specificity #MachineLearning #prediction challenge is live now! Spread the word! @airr_community @iRecRepPapers
https://t.co/lDMxM2jOW0
kaggle.com
Competitors will make predictions on previously unpublished TCR-epitope binding data in order to benchmark prediction methods.
1
23
44
(1/8)🎉A fresh preprint in which we present LIgO — a powerful tool to simulate adaptive immune receptor (AIR) and repertoire (AIRR) data for the development and benchmarking of AIRR-based ML 🧵⬇️ https://t.co/0iif0WbUP2
3
21
56
https://t.co/duAWlQXQeC. A good reminder that predictions of TCRs recognizing 'unseen' epitopes (i.e., epitopes without any known TCR) remains an unsolved challenge.
biorxiv.org
The ability to predict binding between peptides presented by the Major Histocompatibility Complex (MHC) class I molecules and T-cell receptors (TCR) is of great interest in areas of vaccine develop...
1
10
38
8/8 We thank @HelmsleyTrust, @forskningsradet, @UniOslo, @UiO_LifeSci, @EU_H2020, @ireceptor_plus, Norwegian Cancer Society, @JCoDiRC, Innovative Medicines Initiative 2 Joint Undertaking for generous funding.
0
0
3
7/8 Huge thanks to Ludvig M. Sollid, @SandveGeir, and the co-last authors @PRobertImmodels and @victorgreiff
1
0
3
6/8 With our model AIRRTM, we show that in some settings it is possible to identify sequences responsible for the repertoire labels and even generate novel sequences with the same property!
1
0
3
5/8 Can one use only repertoire labels – and thus utilize large-scale AIRR-seq data – to make predictions for individual sequences? Well, in some settings you definitely can!
1
0
3
4/8 At the same time, for ML on repertoires, one only needs repertoire labels, usually available from patient metadata stored in public databases.
1
0
3
3/8 ML on individual sequences arguably has more impact on drug design and discovery – but it requires data which are expensive to produce (e.g. display technologies).
1
0
3
2/8 Machine learning (ML) on adaptive immune receptor repertoires can be done on individual sequences (e.g., predict TCR specificity or generate antigen-specific antibodies) or on whole repertoires (immunome-based diagnostics).
1
0
3
1/8 New preprint: generative modeling of AIRR repertoires, the last piece of my PhD, >2 years of work, a project that is very dear to me https://t.co/gwZt4WBuLY
2
9
44
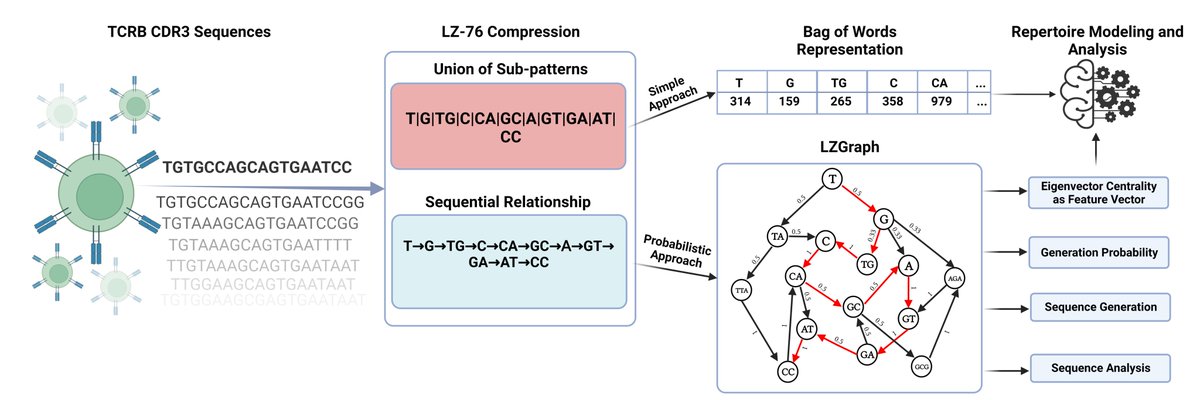
1/5 🎉 Thrilled to announce our paper on a novel approach to T-cell receptor beta chain (TCRB) repertoire encoding using lossless string compression has been published in Bioinformatics! This work is pivotal for our daily work and analysis of TCRB's #Bioinformatics #Immunology
1
23
119
Excited to present the Hoehn Lab @GeiselMed website! Opening doors this September. Already have a postdoc position open - please get in touch if interested in B cell phylogenetics with single cell data. More to come soon.
3
10
27
Efficient evolution of human antibodies from general protein language models https://t.co/F4YxXyMZw8
3
62
171
(1/n) A fresh preprint in which we present a novel approach to T-Cell Receptor Beta chain (TCRB) repertoire encoding using lossless string compression. https://t.co/gtLaPcZFbk
2
20
55
IGHV allele similarity clustering improves genotype inference from adaptive immune receptor repertoire sequencing data https://t.co/Tunw4KACeB
#biorxiv_immuno
biorxiv.org
In adaptive immune receptor repertoire analysis, determining the germline variable (V) allele associated with each T- and B-cell receptor sequence is a crucial step. This process is highly impacted...
0
3
10