Piotr Draczkowski
@PDraczkowski
Followers
32
Following
108
Media
1
Statuses
42
Bioinformatician / Research engineer in structural biology @SciLifeLab. Outdoor addicted. Old-fashioned analogue photography enthusiast.
Joined February 2013
📣 Job alert! Our partner group, the Dioscuri Centre for Modeling of Posttranslational Modifications @BioPhysMatt, is seeking a talented individual in the field of #ComputationalBiophysics. Apply ✏️✅! #AssistantProfessor 👉 https://t.co/wP0FP4MJ3o
0
2
6
Very pleased to know that our recent work on hCoV-229E S:hAPN recognition has been highlighted by Nature Communications alongside the work of the newly minted Nobel Prize laureate David Baker
lnkd.in
This link will take you to a page that’s not on LinkedIn
0
1
6
ESMAdam: a plug-and-play all-purpose protein ensemble generator 1. ESMAdam introduces a versatile framework for protein ensemble generation, bridging gaps in high-throughput applications. Leveraging the pretrained ESMFold model and Adam optimization, it excels in efficiency and
0
21
84
This is exciting and opens new avenues for studying the proteasome-ubiquitin system. I am now awaiting K11 ubiquitin-specific probes ;)
pubs.acs.org
Intracellular monitoring of protein ubiquitination and differentiating polyubiquitin chain topology are crucial for understanding life processes and drug discovery, which is challenged by the high...
0
0
2
Here’s a brief teaser for our latest preprint reporting the first structures of the K11/K48-branched Ub chain bound to the human proteasome: https://t.co/illzy1JT4X Check the full text for more details: https://t.co/D9BMaW7poF Thx to @asibc512 for making this possible!
biorxiv.org
Beyond the canonical K48-linked homotypic polyubiquitination for proteasome-targeted proteolysis, K11/K48-branched ubiquitin (Ub) chains are involved in fast-tracking protein turnover during cell...
0
1
2
Our preprint on the first glimpse of how the human proteasome recognizes a K11/K48-branched ubiquitin chain is out. This work reveals a new multivalent ubiquitin binding mode by the proteasome that could serve as priority checkin signal for proteolysis. https://t.co/Sk2ERBDb9B
1
9
13
Call for AMBER is now open and closes on September 6th. Prof. @A_Chacinska is seeking 2 postdoctoral researchers for 2 projects in structural biology, and molecular cell biology and proteomics. Link to the project website: https://t.co/DsUWs1LSeJ
@FNP_org_pl @PAN_akademia
0
5
8
How proteins are imported into human mitochondria? Read the latest publication from @A_Chacinska lab. Find out how Klaudia Maruszczak and @PDraczkowski used computational tools to get first architectural insights into these processes. @NCN_PL
1
2
8
Everyone (including me :) is hyped up about the new AF3 today, so let’s mix it up with some other cool science updates from yesterday:
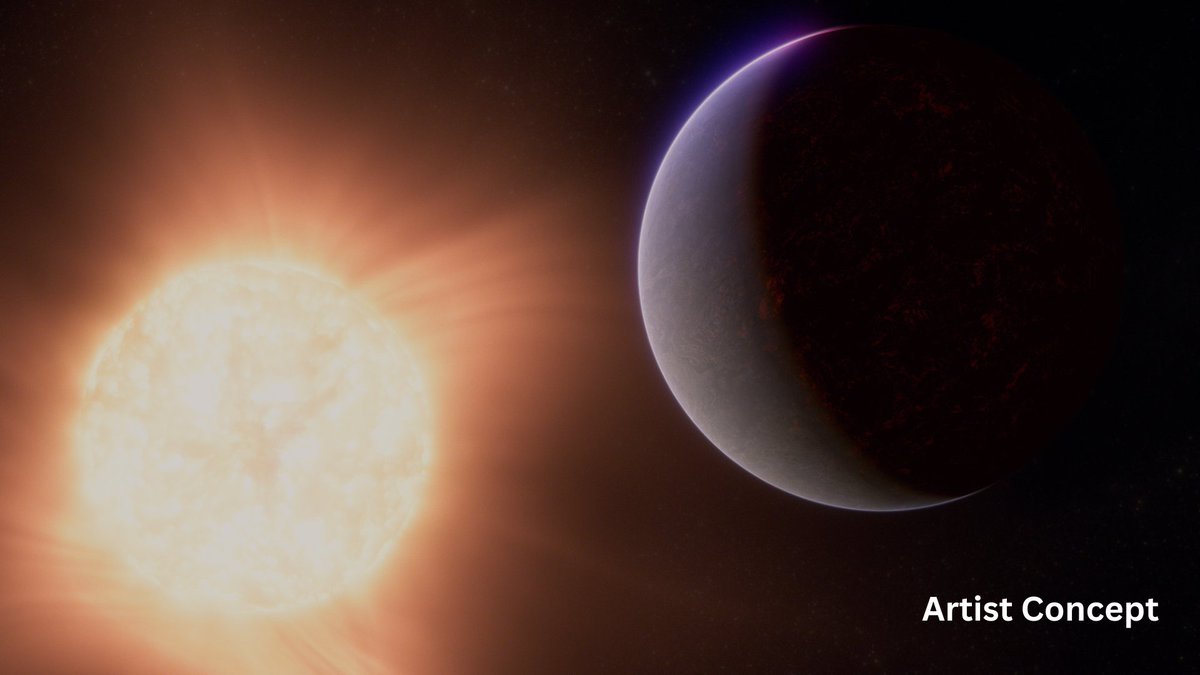
The floor is lava! 🔥 Webb may have detected atmospheric gasses around molten 55 Cancri e, 41 light years from Earth. It’s the best evidence to date for a rocky planet with an atmosphere outside our solar system! https://t.co/8pl9VAIS1M
0
0
3
Love it :D @wojtek_pokrzywa have an idea to level up the difficulty: how about adding branched K11/K48 Ub chains into the game scenario? ;D
Check out our game, DEGRADATOR! 🎮 Designed to popularize and educate on the ubiquitin-proteasome system and PROTACs. Perfect for kids 12+ but not only 😉 Dive into 10 fun levels to explore protein degradation. Available on Google Play and https://t.co/TMiwcfLi9M 🚀
0
0
4
If you want a brief intro to the latest structural discoveries in mitochondrial protein import, check out our piece in @NatureSMB. Thanks to @AgnieszkaC59642, @MichWasil and whole brilliant team @IMol_Institute for the exciting collaboration. https://t.co/y9Mga4teQi
nature.com
Nature Structural & Molecular Biology - The mitochondrial translocase complex TIM23 targets several hundreds of proteins to their location in the mitochondrial matrix or inner membrane. Recent...
2
5
24
A lot is happening at the interface of AI and cryoEM. It's great to see another method aiming at resolving structural heterogeneity of the sample as this is where the cryoEM really shines.
1/ Very excited to present cryoSTAR, a novel approach for continuous heterogeneity in SPA cryo-EM. In brief, cryoSTAR leverages the prior knowledge from a user-given atomic model to better find dynamics in the final reconstruction. https://t.co/WaaTUnMQRM
https://t.co/2SDzNSjPEs
0
0
3
1/ Very excited to present cryoSTAR, a novel approach for continuous heterogeneity in SPA cryo-EM. In brief, cryoSTAR leverages the prior knowledge from a user-given atomic model to better find dynamics in the final reconstruction. https://t.co/WaaTUnMQRM
https://t.co/2SDzNSjPEs
6
43
177
👇🏽 of outstanding importance 👇🏽
Jane S. Richardson et al.: The bad and the good of trends in model building and refinement for sparse-data regions: pernicious forms of overfitting versus good new tools and predictions #CaBLAMValidation ... #IUCr
0
2
10
New! We’ve just put up a note evaluating the latest, in-development version of AlphaFold (“AlphaFold-latest”). This is a preview - development is still in progress - but performance across a wide range of tasks is striking. https://t.co/28nuVOir9v Highlights in the thread. 1/7
8
247
800
Realizacja zapowiedzi zmian w finansowaniu badań będzie ogromnym ciosem dla nauki (...) Dlatego apelujemy o zachowanie niezależnego i opartego na ocenie eksperckiej sposobu działania @NCN_PL – piszą laureaci grantów #ERC w liście do władz państwa https://t.co/Yu6Bskudoe
0
30
76
IMol director, Prof. Agnieszka Chacińska, has been ranked among the world's best scientists in Biology and Biochemistry area. Congratulations! source: https://t.co/nNiVGDwNb6 Leading Academic Research Portal https://t.co/hFFr7iiwB3…
@FNP_org_pl
@NCN_PL
@PAN_akademia
0
1
8
So does everyone's desktop just look like this after making figures in @UCSFChimeraX , or is this just me? #Cursed #CryoEM (changing the working directory doesn't save them there 🤷♀️)
9
4
30