Structural Biology
@ActaCrystD
Followers
3K
Following
1K
Media
496
Statuses
5K
Acta Crystallographica Section D: Structural Biology is a leading structural biology journal published by the IUCr.
Chester, England
Joined May 2012
Acta Crystallographica Section D - Structural Biology welcomes the submission of articles covering all aspects of structural biology, with a particular emphasis on the structures of biological macromolecules or the methods used to determine them. ➡️
0
4
9
A set of 229 crystal structures of human and mouse FABPs are reported with a median resolution of 1.2 Å, 216 of which have a ligand bound. Many structures also have additional affinity data. #IsoformSpecificity #CrystalPathologies #LigandBinding
0
0
5
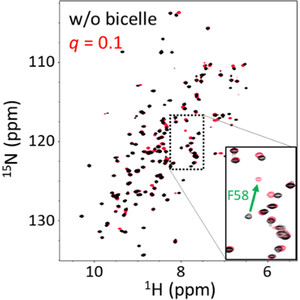
The influence of fatty acids and inhibitors on the conformational flexibility and bicelle/membrane association of human FABP4 was studied using NMR and a preparation completely free of endogenous ligands #NMRDynamics #MicelleAndBicelleBinding
0
1
3
RT @MikeGlazer1: For lovers of #crystallography: The latest issue of the @IUCr Newsletter is out at .
0
7
0
A comprehensive statistical analysis of how Q-scores are related to reported resolution, based on ∼10 000 EMDB maps and associated PDB atomic coordinate models archived in the EMDB @ucsandiego @ActaCrystD @IUCr #CryoEM #QScores #Validation
0
7
21
How can you find consistent biological 3D structures in heavily contaminated electron microscopy data sets without using any prior knowledge? @ActaCrystD @IUCr #CryoEM #SingleParticle #Reconstruction
0
4
13
RT @PracticalFrag: "Unexpectedly high occurrence of wrong ligands." Roche team examines > 200 high quality ligand-bound structures for fat….
0
2
0
Liqing Huang et al.: XFBLD-Platform: a crystallographic fragment-screening platform at Shanghai Synchrotron Radiation Facility #FragmentBasedLeadDiscovery #ProteinCrystallography #FragmentScreening . #IUCr
journals.iucr.org
A crystallographic fragment-screening platform, referred to as the XFBLD-Platform, developed at Shanghai Synchrotron Radiation Facility accelerates fragment-based lead discovery through an integrated...
0
2
7
Elspeth Garman highlights a triumvirate of papers from the Lead Discovery Department at F. Hoffmann-La Roche Ltd in Basel, Switzerland presenting a detailed multi-faceted study of fatty acid-binding protein (FABP) structures @ActaCrystD @IUCr
0
0
3
RT @ActaCrystD: The August issue is out! On the cover: ~15% of the ligands in a high-resolution set of FABP struct….
0
1
0
Rao and Soriano Garcia: Rengachary Parthasarathy (1939–2025): a life journey from Nagari to San Jose . #IUCr
journals.iucr.org
Rengachary Parthasarathy is remembered.
0
0
0
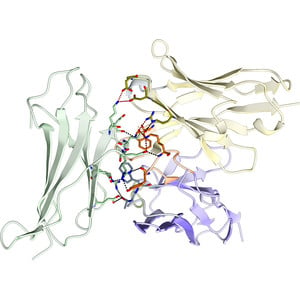
Wu, Zeng, Wang and Wei: Crystal structure of the β2-microglobulin–BBM.1 antibody complex reveals the molecular basis of antigen recognition #Beta2Microglobulin #Antibody #SingleChainVariableFragment . #IUCr
journals.iucr.org
This study presents the crystal structure of human β2-microglobulin (β2M) complexed with the monoclonal antibody BBM.1, revealing critical interactions involving key epitope residues. These structu...
0
0
1
RT @IUCr: Cryo-electron microscopy is now essential in structural biology—transforming how we determine macromolecular structures at near-a….
0
4
0
John R. Helliwell: Time-Resolved Methods in Structural Biology, Methods in Enzymology Volume 709. Edited by Peter Moody and Hanna Kwon. Academic Press, New York, 2024, pp. 412. ISBN-10 044331456X, ISBN-13 978-0443314568. #IUCr.
journals.iucr.org
John Helliwell provides a review of the book Time-Resolved Methods in Structural Biology, edited by Peter Moody and Hanna Kwon.
0
3
10
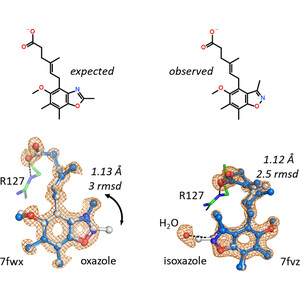
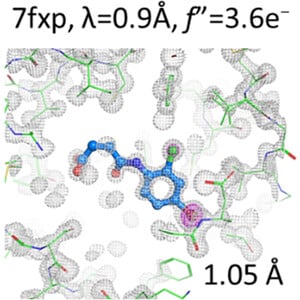
Andreas Ehler et al.: A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands #StructureBasedDrugDesign #FattyAcidBindingProteins #LigandBinding #IUCr.
journals.iucr.org
Very high-resolution fatty acid-binding protein structures identified a substantial number of ligands that differ strongly from their expected structure. If different ligand chemistry is a potentia...
0
0
0
Andreas Ehler et al. A high-resolution data set of fatty acid-binding protein structures. II. Crystallographic overview, ligand classes and binding pose #IsoformSpecificity #CrystalPathologies #LigandBinding #IUCr.
journals.iucr.org
A high-quality set of 229 crystal structures of various isoforms of human and mouse fatty acid-binding proteins in apo and ligand-bound forms is presented. Many ligands have associated affinity data,...
0
0
1
Fabio Casagrande et al.: A high-resolution data set of fatty acid-binding protein structures. I. Dynamics of FABP4 and ligand binding #NMRDynamics #MicelleAndBicelleBinding #ResidualLigands . #IUCr
journals.iucr.org
NMR studies on human FABP4 in solution and bound to micelles and bicelles delineate its dynamics in a ligand-dependent fashion. Residual ligands are co-purified with the protein, which may negatively...
0
0
1
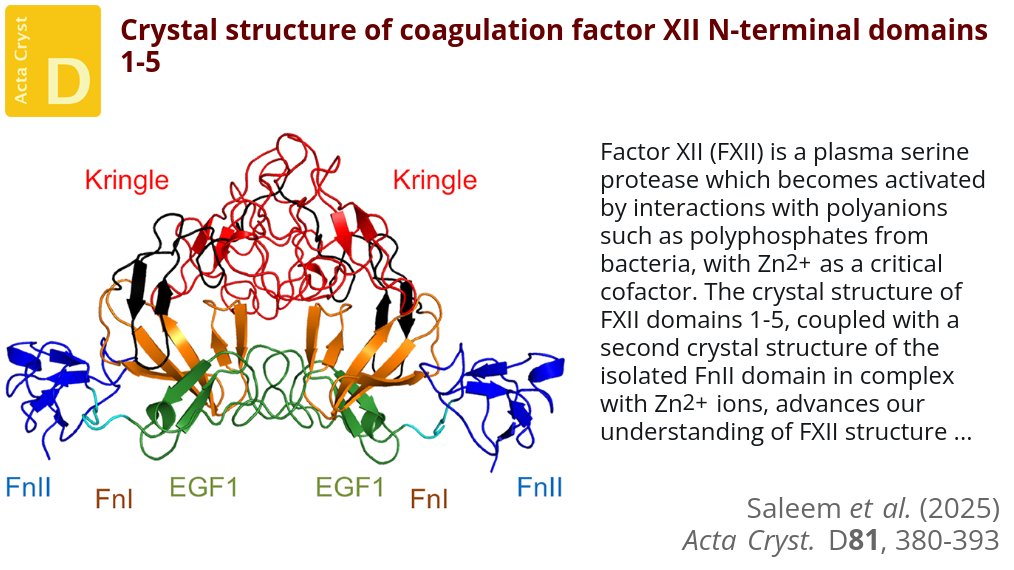
To investigate coagulation factor XII ligand binding, crystal structures of its five N-terminal domains and of its FnII domain bound to Zn2+ were determined @UniofNottingham @universityleeds #FactorXII #FibronectinTypeIIDomain #KringleDomain
0
0
2