Ljubisa Miskovic
@Misko_L34
Followers
32
Following
14
Media
0
Statuses
43
This work by Toumpe, Masid, Hatzimanikatis & Miskovic (@epfl_en LCSB) provides a resource for exploring metabolic regulation, vulnerabilities, and precision oncology. All models are openly available. #systemsbiology #cancermetabolism #multiomics #precisiononcology #openscience
0
1
0
By integrating multi-omics data with enzyme kinetics, we generated a population of near–genome-scale kinetic models that capture BRCA1-mutant vs wild-type ovarian cancer physiology. These models reveal network-wide control principles & drug-response dynamics.
1
1
0
🚀 New preprint from @epfl_en’s Laboratory of Computational Systems Biotechnology (LCSB)! We present “Multi-omics–driven kinetic modeling reveals metabolic vulnerabilities and differential drug-response dynamics in ovarian cancer.” 🔗 https://t.co/EaqYTn8bRq
1
1
0
Our review made the cover in ACS Synthetic Biology! 🎉🧬 We explore how large-scale kinetic models are shaping the future of metabolic engineering — check it out here 👉 https://t.co/GsO6bv8JTF Big thanks to the team and @ACSPublications! #MyACSCover #SyntheticBiology
pubs.acs.org
Recent advances in mechanistic kinetic modeling now enable detailed descriptions of metabolism across the universe of living organisms. We highlight the potential of these developments to drive...
0
2
3
Excited to share our new preprint, where we present NIS, a framework that distills GEMs into interpretable modules, enabling direct cross-species comparisons of fueling pathways, biosynthesis, and environmental exchanges. Congratulations to the authors!
👏 A big thank you to my co-authors Meric Ataman & Vassily Hatzimanikatis for this great collaboration! 🔍 Read more:
0
1
2
This preprint https://t.co/n2RsrifKp7 proposes a systematic framework to repurpose generative neural nets across physiological contexts enabling efficient construction of kinetic models tailored to specific scenarios. A step toward flexible/scalable modeling! #AI #SystemsBiology
biorxiv.org
Generative machine learning methods that use neural networks to parameterize large-scale and near genome-scale kinetic models have delivered significant efficiency gains in model construction, paving...
0
0
0
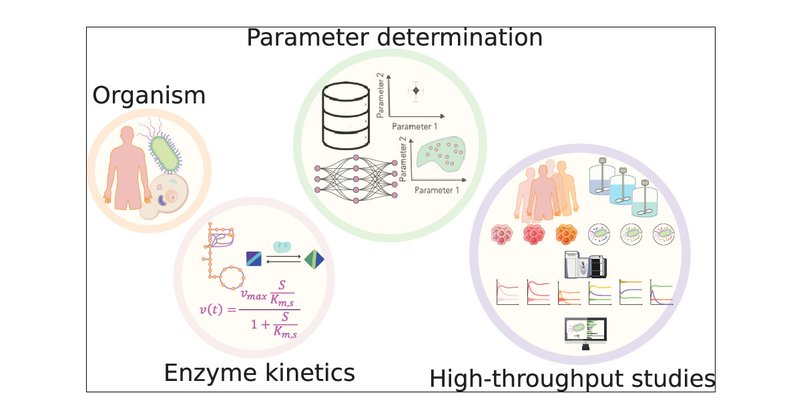
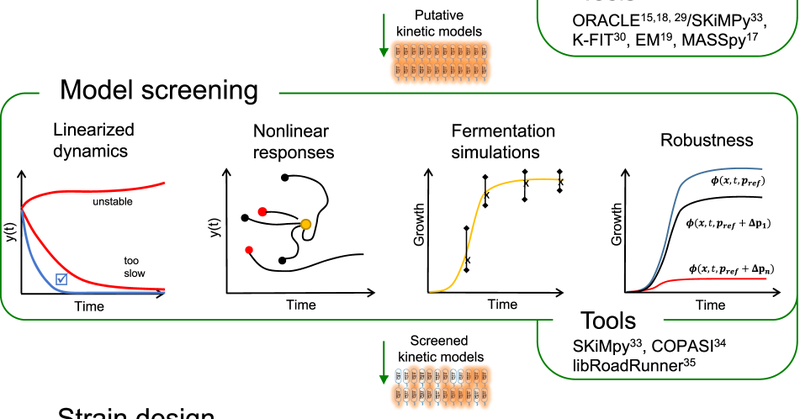
The Dawn of High-Throughput and Genome-Scale Kinetic Modeling: Recent Advances and Future Directions #machinelearning #compchem
pubs.acs.org
Researchers have invested much effort into developing kinetic models due to their ability to capture dynamic behaviors, transient states, and regulatory mechanisms of metabolism, providing a detailed...
1
4
9
This work showcases one of the few successful integrations of kinetic modeling and experiments to optimize cell factories. It highlights the power of NOMAD ( https://t.co/2zbJiEMFWi) in accelerating and improving strain design!
nature.com
Nature Communications - No consensus exists on the computationally tractable use of dynamic models for strain design. To tackle this, the authors report a framework,...
0
3
1
Excited to share our preprint on boosting p-coumaric acid production in S. cerevisiae! 🎉 In collaboration with Irina Borodina's group (@Irina__Borodina), we used our NOMAD framework for strain design strategies. Read more:
biorxiv.org
The use of kinetic models of metabolism in design-build-learn-test cycles is limited despite their potential to guide and accelerate the optimization of cell factories. This is primarily due to...
1
2
1
Want to efficiently create large-scale dynamic models of metabolism that fit experimental data and reliably predict metabolic responses to various perturbations? Our new method does just that! Check it out here: https://t.co/d757bhiFC8.
nature.com
Nature Catalysis - Despite the availability of large omics datasets, determining intracellular metabolic states is challenging. Now a generative machine learning framework called RENAISSANCE has...
0
1
6
Congratulations to Subham (@astro_dank), Bharath (@bharathnarayana), Michael (@moret1788), and Vassily (@Vassily_13)!
0
0
1
Remember RENAISSANCE ( https://t.co/tOe3IRSnP1)? This generative ML method for constructing large-scale dynamic models of metabolism has been accepted to @NatureCatalysis
biorxiv.org
Generating large omics datasets has become routine practice to gain insights into cellular processes, yet deciphering such massive datasets and determining intracellular metabolic states remains...
1
4
4
Our recent publication in @NatureComms, “Rational strain design with minimal phenotype perturbation” by Narayanan et al., has been showcased in the recent Editors’ Highlights for Biotechnology and Methods focus ( https://t.co/YLmSKMX85k).
1
3
5
Can we efficiently use dynamic nonlinear models to devise genetic interventions for desired cellular phenotypes? Check out in https://t.co/2zbJiEM86K just published in @NatureComms. Congratulations to Bharath (@bharathnarayana), Daniel (@realDRWeilandt), Maria, and @Vassily_13.
nature.com
Nature Communications - No consensus exists on the computationally tractable use of dynamic models for strain design. To tackle this, the authors report a framework,...
0
0
1
Curious and want to dive deeper? Check out our preprint on Bioarxiv:
biorxiv.org
Novel sequencing techniques and biochemical pathway prediction resources provide a wealth of data on novel proteins and computationally predicted enzymatic reactions. Accurate matching of protein...
1
1
1
Excited to announce that our latest work in now online ! This time the team significantly expanded on their previous tool, BridgIT, an enzyme annotation for orphan and novel reactions, to bring you the new and improved BridgIT+ ! Read the thread below to find out more :)
1
2
3
Our latest work on simulating plasmid burden using ME-models is now available at https://t.co/5wihMd8v00! Congratulations to Omids (@oftadehomid) and Vassily (@vassily_13)!
biorxiv.org
The production of recombinant proteins in a host using synthetic constructs such as plasmids comes at the cost of detrimental effects such as reduced growth, energetic inefficiencies, and other...
Finally out! In this paper, we used models of metabolism and expression (ME-models) to simulate the plasmid burden. We found the optimal plasmid copy number to achieve the best trade-off between biomass and product yield. https://t.co/KLxKnSVo0i
0
1
8
Ever thought about how evolution shapes enzyme binding mechanisms? Exited to share this work of @aslisahin2205 @realLCSB with @Vassily_13. We show that cellular concentrations and thermodynamics determine the optimal binding sequence and enzyme saturation.
0
0
1
Really happy to have this preprint out! Quick thread about our work:
very proud of the collaborative work with @MumenthalerLab @MathCancer @ngraham to study metabolic crosstalk! and @GelbachPatrick has a new @biorxivpreprint presenting metabolic models of macrophages in #ColorectalCancer. check it out:
1
2
7
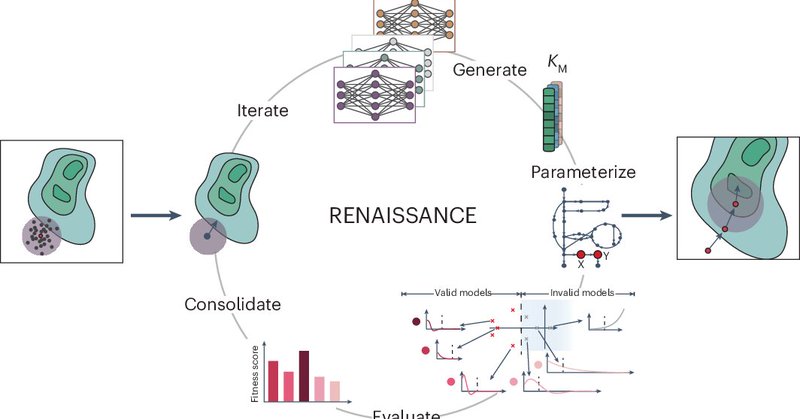
RENAISSANCE does not require training data to parameterize nonlinear dynamic models of metabolism. Instead, it uses evolution strategies, thus allowing specialists and non-specialists to efficiently create large-scale kinetic models. How? Check out:
biorxiv.org
Large omics datasets are nowadays routinely generated to provide insights into cellular processes. Nevertheless, making sense of omics data and determining intracellular metabolic states remains...
0
3
9