EichlerLab
@EichlerLab
Followers
1K
Following
133
Media
18
Statuses
75
Eichler Lab @UW Twitter @uwgenome #hhmi
Joined April 2022
Check out new AoU collaboration paper: Long-read sequencing of 1027 All of Us participants self-identified as African American discovers new structural variant disease associations: https://t.co/04nSZ5Ok1D
@EichlerEE @uwgenome @KiranGarimella
medrxiv.org
The All of Us Research Program (AoU) is a national biobank seeking to enroll one million individuals in the United States to link genomic and biomedical data, including short- and long-read whole-g...
0
6
17
Thank you, Dr. Danny Miller, for hosting, and fantastic job, Dr. @fkmastrorosa! https://t.co/yfTkbe081e
brotmanbaty.org
BBI
0
1
2
Recent PhD grad Dishuck unveiled NPIP structural variation & evolutionary dynamics across 169 human haplotypes—revealing brain-expressed paralogs, selective sweeps & neofunctionalization. 🎧 @BasebyBasePod Ep. 129: Dives into NPIP’s role in our genome: https://t.co/rFwzJ5S9pW
1
8
24
🧬 How fast do humans mutate? Our groundbreaking study tracks DNA changes across 4 generations to reveal the pace of human evolution revealing insights into our genetic legacy. Watch now 👉 https://t.co/SazD16ZDum
#Genetics #Evolution #ScienceVideo #Nature
0
2
11
Check out our postdoc's updated preprint!
We updated our preprint describing chr21 centromere genetics/epigenetics in families with Down syndrome and general population! We found transgenerational methylation changes in a subset of families and that centromere size asymmetry is exclusive to T21! https://t.co/nLdsUzClyE
0
0
3
Using >130 human & 12 primate haplotypes, we reconstruct the chromosome 22q11.2 evolution to identify haplotype structures linked to deletions or inversions, explaining the lower prevalence of 22q11.2 deletion syndrome in individuals of African descent. https://t.co/07Yum0L3hc
1
22
69
The Eichler lab's new, revamped website is now live! Learn more about the lab's research and scientists @ https://t.co/U0TlNtB3ov
0
3
20
Always fun to see the "band" get back together! Cheers to former lab members!
@devin_locke Next, pangenome-based segmental duplication discovery and phasing with the boss @EichlerLab w/ @DavisCompGen and @hajirasouliha
0
3
14
Our Nature paper ( https://t.co/8ucXysP2w8) deep sequencing a 4-generation, 28-member family using multiple sequencing technologies to study transmission of all classes of genetic variation is out! @EichlerEE @uwgenome @HHMINEWS
Advanced genomic analysis of 4 generation family offers new knowledge about genetic mutations & their transmission, including inherited variants & those that arise anew @Nature @uwgenome @EichlerLab @UWMedicine @UUtah @PacBio
https://t.co/DRYYOenRZm
2
22
60
Check it out! Telomere-to-telomere ape genome assemblies reveal new biology & insights into evolution: https://t.co/Xxy8flYHd2 Nature News: https://t.co/XGqArEaiaK Nature Podcast: https://t.co/TsLokU0d83
@DongahnYoo @EichlerEE @uwgenome @HHMINEWS
nature.com
Nature - Hear the biggest stories from the world of science | 09 April 2025
Polished genomes of 6 ape species - siamang, Borneo & Sumatran orangutan, gorilla, chimp & bonobo - have been assembled & comparatively analyzed. Findings shed new light on human & ape evolution @Nature @Eichlerlab @uwgenome @UWMedicine
https://t.co/dp4glyZPcM
0
7
17
Check out this ASHG podcast episode with our grad student Lizzie Plender! She talks about how she got into genetics and why mucins are the "coolest" genes out there: https://t.co/lygnR6xpOn
https://t.co/270Wtfra6K
0
1
10
Our new Nature Genetics paper on human segmental duplications (SDs) provides a pangenome perspective of SDs, new potential protein-coding genes, and greater complexity in human variation with implications for disease and evolution. https://t.co/CYCI8xQEiH
0
34
129
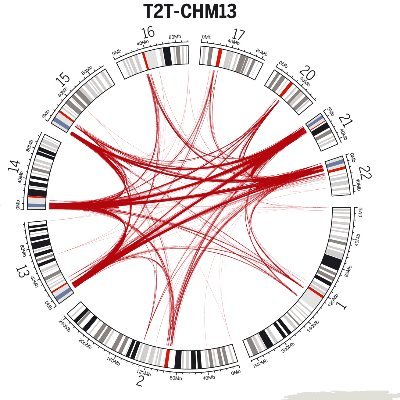
I'm thrilled to share our latest research on the human chromosome 2 fusion, addressing a basic question: "What makes us uniquely human?". Our work is now live on BioRxiv! ( https://t.co/qA2etlHQKf).
@EichlerLab We’d love to hear your thoughts and feedback! :)
biorxiv.org
All great apes differ karyotypically from humans due to the fusion of chromosomes 2a and 2b, resulting in human chromosome 2. Yet, the structure, function, and evolutionary history of the genomic...
1
7
39
Congrats Dr. Philip Dishuck @genesofplenty on your successful "Structural variation and expression of segmentally duplicated human genes" dissertation defense. Your determination and resilience have paid off! To your future success! Cheers!!
1
0
13
Eichler lab grad student Xavi Guitart (@xavster838)'s first author paper "Independent expansion, selection, and hypervariability of the TBC1D3 gene family in humans" featured in a GR special issue! @EichlerEE @uwgenome @HHMINEWS
SPECIAL ISSUE! This month @genomeresearch publishes a diverse collection of research and review articles in a special issue highlighting advances in long-read sequencing applications in biology and medicine. https://t.co/4ezPHyvmXH.
0
5
16
We're hiring! Join our team working with cutting-edge genomic technologies. Check out our new long-read sequencing tech position @ UW in Seattle: https://t.co/4h1HgQytT0
#Genomics #UW #PacBio
1
17
31
Congrats Dr. Michelle Noyes @mitch_does_sci! You conquered the academic battlefield w/ your brilliant defense: a masterclass in dedication, knowledge, & eloquence w/ your unique quirkiness. This milestone is a testament to your hard work & perseverance. To your future success!
0
1
32
Check out SVbyEye: A new visualization tool to characterize structural variation among whole-genome assemblies ( https://t.co/G0zGoHxZpe)
0
78
221
Now on bioRxiv! Deep sequencing of a four-generation, 28-member family (CEPH 1463) using multiple sequencing technologies provides an important resource to study transmission of all classes of genetic variation. https://t.co/fRxtUCirOr
0
30
67








