Shouchao Yan
@KedderSc
Followers
122
Following
732
Media
3
Statuses
146
JIF Rotation PhD Student interested in the biosynthesis and functions of legume natural products | Metabolomics| Osbourn group at John Innes Centre 🌱🫘
Norwich, England
Joined April 2021
RT @BeniddirM: It is with tremendous emotion that I share with you a community effort @jcheminf that resulted in t….
0
6
0
RT @Pdorrestein1: This preprint allows for “explanation” of the ion forms using khipu ore annotation and conclude that the dark matter are….
0
8
0
RT @jenny_sjo: 🌸🧼We made the cover⚕️🌱. 📢Check out the February issue of @nchembio✨️. The cover illustration is an antique soapwort drawing….
0
13
0
RT @JohnInnesCentre: PRESS RELEASE - Could this fundamental discovery revolutionise fertiliser use in farming? . Researchers have discovere….
0
22
0
RT @YazhongXigua: Breakthrough in crops! 🌱 🔥💥.Using prime editing, Xu Lab engineered heat-responsive genes to optimize source-sink relation….
0
72
0
RT @Chemistophe: We are excited to share our work on the detoxification of solanidine, a steroidal alkaloid (StA) toxin☠️found in potato🥔,….
0
14
0
RT @TrendsPlantSci: Multiple layers of regulators emerge in the network controlling lateral root organogenesis http….
0
44
0
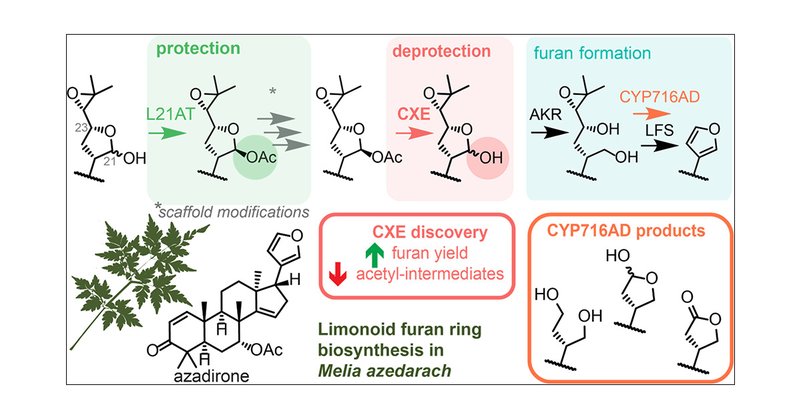
RT @HannahHodgson93: Delighted that our latest limonoid biosynthetic pathway work is now online @J_A_C_S !. We characterise a yield boostin….
pubs.acs.org
The furan ring is a defining feature of limonoids, a class of highly rearranged and bioactive plant tetranortriterpenoids. We recently reported an apparent complete biosynthetic pathway to these...
0
17
0
RT @nobolly: Reverse engineering of the pattern recognition receptor FLS2 reveals key design principles of broader recognition spectra agai….
biorxiv.org
In the ongoing plant-pathogen arms race, plants employ pattern recognition receptors (PRRs) to recognize pathogen-associated molecular patterns (PAMPs), while in successful pathogens, PAMPs can...
0
11
0
Congratulations!!!!.
#ProudPI in #CompMetabolomics - really great to see how this project came to fruition over the last 1.5 years - many congratulations 👏 🎉 to Kevin!! Thanks to @WU_BioInfo, @WURplant, @WUR for resources and funding! 😎 #metabolomics.
0
0
2
RT @BeniddirM: So proud to share our recent chemical standpoint of emerging trends in plant natural products biosynthesis: .
0
12
0
Congratssssss!!!!! I’m so happy for you!!!!!! ❤️❤️❤️.
I feel excited and honoured that @ERC_Research selected my project for funding! 🎉. With our #ERCStG "FRIENEMIES" we will investigate the #evolution of plant-fungus interactions from pathogenic to beneficial in land plants @ZMBP_Tuebingen @uni_tue. #EvoMPMI #PlantSci.
1
1
2
RT @LauraLSaavedra: Great work about the emergence of the CYP73 gene family as a foundational event in the development of the plant phenylp….
0
8
0
RT @mingxunwang: I am very proud of a first year PhD student in my lab, Xianghu Wang, for his paper on how to improve molecular networking….
0
14
0
RT @HongyanZhu1: Excited to share our new publication: Species-specific microsymbiont discrimination mediated by a Medicago receptor kinase….
science.org
A plant receptor–like kinase enacts partner specificity between Medicago and two closely related rhizobial species.
0
22
0
🙌🙌🙌🙌🙌🙌🙌.
Thanks to @ThePlantJournal for this interview and especially to @Luis_deLunav for making it so easy!.If you want to know more about being a #JuniorPI @ZMBP_Tuebingen, check this 👇.
0
0
2