Justin van der Hooft
@jjjvanderhooft
Followers
6K
Following
39K
Media
2K
Statuses
16K
Assistant Professor in Computational Metabolomics @vdHooft_CompMet @WU_BioInfo @WURplant @WUR! 😎 | @wyoungacademy | #OpenScience! | Visiting Professor @go2uj
Joined November 2013
Proud of our team! 😊 I look forward to continuing pushing boundaries in Computational Metabolomics and Integrative Omics together! 💪 #metabolomics #team #community #ProudPI in #CompMetabolomics #naturalproducts #discovery
Our team this month and we were nearly complete!😎#CompMetabolomics #Metabolomics #Wageningen @WU_BioInfo @WURplant @WUR
1
1
51
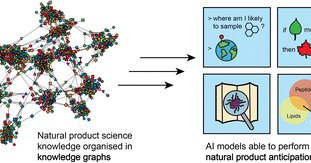
🔓Don't miss this #OpenAccess viewpoint article by @jjjvanderhooft, @marnixmedema, @SkiredjAHub & co @WUR @BioCIS @MIT in our latest issue on leveraging multimodal data and knowledge graphs to empower natural product science with AI Check it out here🔽 https://t.co/njxbSawOWn
pubs.rsc.org
Artificial intelligence (AI) is accelerating how we conduct science, from folding proteins with AlphaFold and summarizing literature findings with large language models, to annotating genomes and...
0
6
11
🔔 PUBLICATION ALERT 🔔 antiSMASH 8.0: extended gene cluster detection capabilities and analyses of chemistry, enzymology & regulation 🦠🧬 Authors from MARBLES: Mitja Zdouc, @jjjvanderhooft, @GillesvanWezel & @marnixmedema (@LeidenBiology @UniLeidenNews & @WU_BioInfo @WUR)o
1
5
13
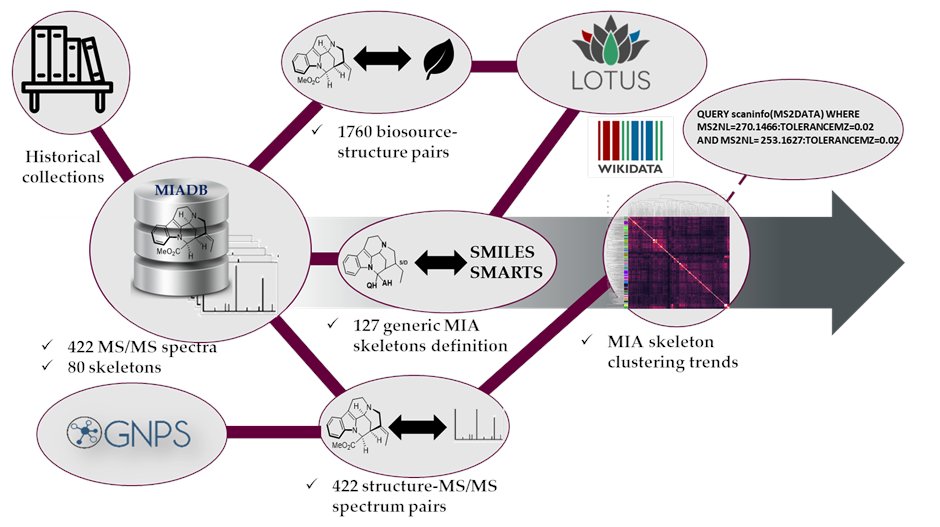
It is with tremendous emotion that I share with you a community effort @jcheminf
https://t.co/sOcWUoi8zx that resulted in the update of the MIADB @GNPS_UCSD and the generation of valuable spectrometric signatures that could be used as queries #MassQL 🙏@Adafede and S. Szwarc
0
1
11
Highlighting new @EMN_MetSoc member, @ellendepaepe, Dr-Assistant and Post-doc @ugent, applying #metabolomics + #lipidomics to study pediatric food allergies, obesity + the #exposome. Ellen contributes to FLEXiGUT, FAME + MeMoSA, w/ a passion for non-invasive biofluids #EMN_MetSoc
8
4
17
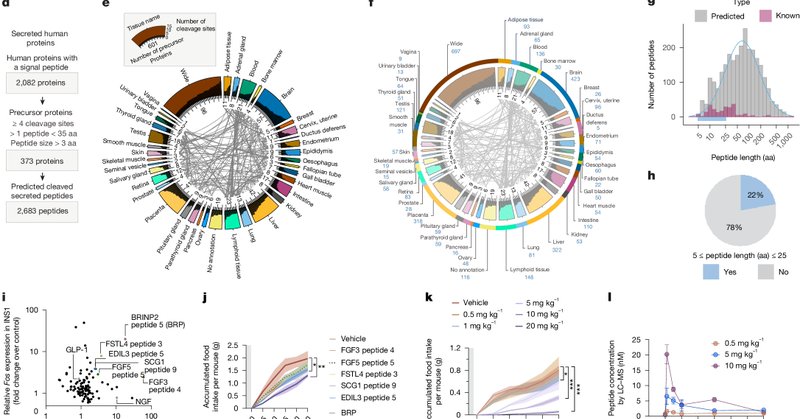
Thrilled to share that my work is out in @Nature. Here, we introduce Peptide Predictor, a tool for identifying new bioactive peptides. Using this method, we discovered BRP, an anti-obesity peptide. Huge thanks to the @SvenssonLab and all our collaborators!
nature.com
Nature - Computational drug discovery is used to identify a 12-mer peptide derived from BRINP2 with potent anti-obesity effects that are independent of leptin, glucagon-like peptide 1 receptor and...
17
123
414
This new piece showcases 100 natural products. Many of their syntheses have had significant impacts in org synth. There are many different structural and functional motifs that have required amazing innovations in synthesis. And one of mine is in there too! #chemtwitter #sciart
1
15
146
Yesterday we attended a great talk by @jjjvanderhooft on Metabolomics and current computational tools used in natural products research. Thank you to @go2uj @UJScience @UJLibrary @fideletu for making it possible ✨ #metabolomics #naturalproducts #metabolome #CompMet
0
3
12
It was an honor to play a small role in this… great to see your vision on this topic out in this review, @GarimaSingh_GS !
Our review article exploring how recent research advances in #genomemining, #chemoinformatics, #phylogenies can be combined to unlock the #biosynthetic potential of #lichens - is out now: https://t.co/ncGR6yKUHw. Big thanks to all involved @FDalGrande @fmartin1954 @marnixmedema
1
3
17
Exciting opportunity to join our group as postdoc! Apply now for this project on computational (meta)genome mining as part of a collaborative European research consortium: https://t.co/jJ7qHkYLlv Please share!
4
21
52
Yesterday we had our first insightful workshop with @jjjvanderhooft @go2uj @fideletu . The day was filled with discussions on various computational tools that assist with metabolite annotation in the Metabolomics field. #metabolomics #Metabolome #naturalproducts #microbiome
0
2
8
This preprint allows for “explanation” of the ion forms using khipu ore annotation and conclude that the dark matter are mostly real compounds - still dark. They find less than 10% ISFs. In the end ISF prevalence is dependent on instrument tuning, type/amt of analyte, matrix, etc
“Therefore, the "dark matter" of metabolomics is largely explainable.” in a new paper by Shuzhao Li. https://t.co/rPdiKosolz
https://t.co/SY7tFLRbVG
2
8
28
TTL is excited to be hosting @jjjvanderhooft @vdHooft_CompMet who’s one of our esteemed collaborators and a visiting Professor @go2uj @UJScience . The next couple of days are surely going to be filled with much learning and fun!✨ @fideletu
#Metabolomics #CompMet #metabolome
0
3
11
I am glad to announce that I am one of the 8 winners of the 4th Edition of the #FAIR Data Fund! @4TUResearchData is an international data repository for science, engineering and design. They offer research dataset curation, sharing, long-term access and preservation services.
0
0
8
Great example of metabolites discovered with molecular networking.
Discovery of a family of bile acid-methylcysteamines @Nature @Cornell @group_Schroeder #activitymetabolomics A nuanced interplay between host and microbiota in regulating bile acid signaling pathways through FXR with a new target for metabolic disorders https://t.co/VvnLBbGJ9A
1
13
55
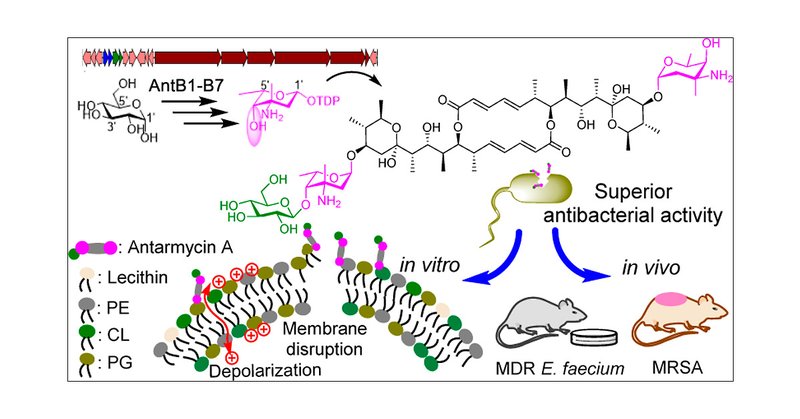
Antarmycins: Discovery, Biosynthesis, Anti-pathogenic Bacterial Activity, and Mechanism of Action from Deep-Sea-Derived Pseudonocardia antarctica | JACS Au
pubs.acs.org
The rapid emergence of antimicrobial-resistant pathogenic microbes has accelerated the search for novel therapeutic agents. Here we report the discovery of antarmycin A (1), an antibiotic containing...
0
10
19
Such a wonderful SIMB workshop on @GNPS_UCSD covering classical mol networking, FBMN, ion identity based mol networking, FBMN-STATS, gnps-dashboard, CMMC-kb, Modifinder, MicrobeMASST, MassQL, and limited ReDU - many were covered at a workshop for the first time. 1/4
3
7
45
new: "MolNexTR: a generalized deep learning model for molecular image recognition" https://t.co/htgNpGUBjy
0
5
16
Good papers lead to more questions than they answer. That's because (as Blaise Pascal wrote) knowledge is like a sphere: the larger it gets the larger also its surface, which is its interface with the unknown.
12
61
376