James Mulqueeney
@JamesMulqueeney
Followers
418
Following
348
Media
24
Statuses
95
Research fellow at the Smithsonian National Museum of Natural History. Former PhD student at University of Southampton and Natural History Museum, London.
Washington, DC
Joined September 2022
Thrilled to share my second PhD paper focused on the application of landmark-free geometric morphometrics! Excited to contribute to advancing shape analysis techniques in biology. Check it out here: "
https://t.co/UTMYBK5b3n"
#PhDResearch #Morphometrics
link.springer.com
BMC Ecology and Evolution - The study of phenotypic evolution has been transformed in recent decades by methods allowing precise quantification of anatomical shape, in particular 3D geometric...
2
29
83
Our paper Regional variability in craniofacial stiffness: a study in normal and Crouzon mice during postnatal development is out in BMMB. This work expands on our previous paper in BMMB. We investigated various loading locations and computational models. https://t.co/InTcacDGYv
0
4
6
Glad to share a new paper led by @yara_haridy and @NeilShubin Lab on the early evolution of teeth! Modern and fossil data point at an ancestrally sensorial function for the earliest dentine https://t.co/hM3Rpa9vQw
nature.com
Nature - Re-examination of the presumed Cambrian fossil fish Anatolepis reveals previous misidentification of aglaspidid sensory structures as dentine, a vertebrate sensory tissue, showing it to be...
1
27
71
Overall, we show the landmark-free methods like Deterministic Atlas Analysis can yield correlated results with traditional manual landmarking & semilandmarking methods. We also propose how to improve these methods in the future!
0
0
3
Finally, we compare the downstream estimates of morphological disparity, evolutionary rates and phylogenetic signal. Here, we find significant correlations between methods for almost all metrics.
1
0
2
We then compare heatmaps of how shape is measured using each method. To do this we compare the shape of different crania with estimated mean shapes of each analysis (landmark top, DAA below). Here is shown for the specimen, Cacajao calvus.
1
0
2
We then compare the correlation between the two methods using Euclidean distances, the Mantel test and the Procrustean randomisation test. We find that using Poisson meshes significantly improves the correlation between methods.
1
0
2
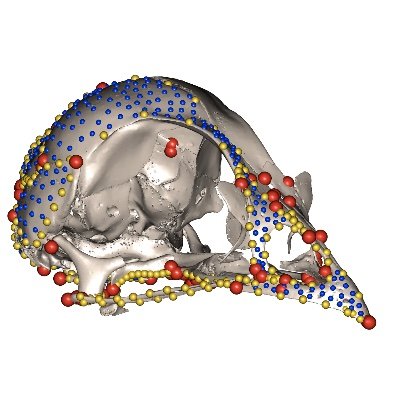
We then visually compare the results of the manual landmarking analysis (754 landmarks and semilandmarks; top left) with the landmark-free approach using 45, 270 and 1,782 control points (top right to bottom left). We notice differences mainly for Cetacea and Primates.
1
0
2
Once overcoming modality issues, we apply the method to 322 placental mammal crania as seen in Goswami et al. 2022 ( https://t.co/3KXpvlRFk5). Here, we apply and test a range of kernel widths, which changes the number of control points used to compare shape (replacing landmarks).
1
0
1
We focus on comparing an implementation of Large Deformation Diffeomorphic Metric Mapping (LDDMM) called a Deterministic Atlas Analysis (DAA) with manual landmarking techniques. We first notice that data modality affects the results, which we overcome making Poisson meshes.
1
0
3
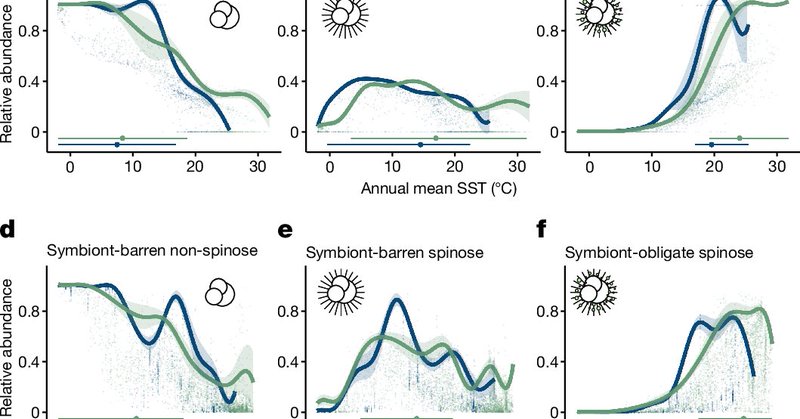
My PhD work on foraminiferal acclimation was published in @Nature. Our study shows that modern warming is threatening planktic foraminifera differently than before. URL:
nature.com
Nature - Data from the fossil record, together with computational modelling, are used to assess the response of foraminifera (marine zooplankton) to temperature changes through time and to predict...
6
13
35
Our final careers panel session of #POP24 on 12 Nov will hear from an Assistant Professor in Polar Modelling, a PhD Researcher, an ESA Research Fellow, and an Evolutionary Biologist, who will explain how we can protect our planet with #maths 🌍 https://t.co/l0LhpTVs8i
#GCW2024
1
2
3
#AI is set to transform the study of #evolutionary #morphology - an interview with Yichen He "One crucial step in using AI effectively is to thoroughly review whether it is suitable for the task at hand..." https://t.co/4LtaR8w8rU
#science #data #biology #morphologist
0
2
2
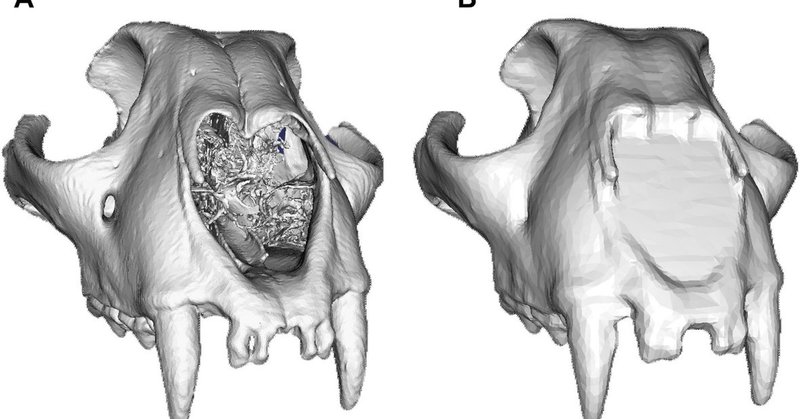
Happy to have had the opportunity to contribute to this special issue and the issue cover! Read the BounTI paper here:
onlinelibrary.wiley.com
BounTI can be used to segment various skeletal features. In this work, we introduced the novel tool and focused on the segmentation of the craniofacial system in various species. While some limitat...
Progress in craniosynostosis research! AdCr meetings unite clinicians, researchers & families to tackle this condition. Latest Journal of Anatomy issue honours Prof. Gillian Morriss-Kay & features 10 interdisciplinary studies. https://t.co/9ZK0FyhpJG
#Craniosynostosis #AdCr
0
3
11
🚨 New Paper 🚨 Our team @NYITAnatomy used #microCT and #GMM to examine how changes in muscle loading affects skull development in chicks. This one is particularly special because it marks the first experimental #embryology study out of my lab 🐣 https://t.co/RO9t80mqZP
0
18
69
Very pleased to share our latest @NHM_Science paper in @RSocPublishing on simulating the formation of trace fossils, with applications for the colonization of land by animals. Led by @ZekunWang3 with Neil Davies, @the_palaeoninja and @nichnos
https://t.co/Ayap8smcTX
0
17
54
Opportunities and Challenges in Applying #AI to #Evolutionary #Morphology Y He , @anjgoswami et al #ArtificialIntelligence (AI) is poised to revolutionize many aspects of #science, including the study of #evolutionary morphology..." https://t.co/Cci6b9aQvz
@anjgoswami @JamesMulqueeney @Emily_C_Watt @geometari @nicole_barber @KnappRew @Scincomancer @ToriHerridge @AgneseLanzetti @gizehrdelazaro @LucyRoberts_NHM Congrats authors. 👏👏👏This paper , however, is from our OA sibling journal @iobjournal We’d love to see it posted over on their account and appreciate all you’re doing to spread the word about this great #science.
0
4
10
Check out this new review paper on the use of AI to study the evolution of morphology. Also this must be a record for most museum students involved in a single publication!
New review paper on using AI to analyse Evolutionary Morphology is officially out! Was great to part of a large collaborative team writing this. https://t.co/2gRSHAxpey
0
1
9
So happy that our paper reviewing current and prospective use of AI in Evolutionary Morphology is out now in @iobopen ! It was great to collaborate with so many wonderful colleagues on this project
New review paper on using AI to analyse Evolutionary Morphology is officially out! Was great to part of a large collaborative team writing this. https://t.co/2gRSHAxpey
1
2
16
Pub 🚨 Review out on the use of AI in evolutionary morphology led by Yichen He, @JamesMulqueeney & Emily Watt! A discussion in @anjgoswami ‘s lab meeting turned into this incredible resource! It’s a massive team effort from Goswami lab & few of us ex- members 🦎 #AI #evomorph
New review paper on using AI to analyse Evolutionary Morphology is officially out! Was great to part of a large collaborative team writing this. https://t.co/2gRSHAxpey
1
2
15