Judith Kribelbauer
@JKribelbauer
Followers
220
Following
827
Media
8
Statuses
106
Gabilan Assistant Professor of Bio Sciences @ USC - Mol&Comp Bio Dept prev @EPFL @Columbia | opinions my own #genomics #mechanisms #TranscriptionFactors
Joined January 2020
Stay tuned, we have some exciting collaborations in the works (@lucapinello lab), applying EXTRA-seq to test cell type specificity & the dynamic range of synthetically created sequences.
1
0
6
We sys compare it to plasmid-based MPRAs and deep learning models (Enformer @deepmind), finding new bio insights & suggesting ways to improve predictions. We resolve expression differences down to distinct TF configurations on DNA, highlighting shortcomings of MPRAs and DL tools.
1
0
3
Finally out! We present EXTRA-seq, a new EXTended Reporter Assay to quantify endogenous enhancer-promoter communication at kb scale!.A 🧵about what it can do:.
biorxiv.org
Precise control of gene expression is essential for cellular function, but the mechanisms by which enhancers communicate with promoters to coordinate this process are not fully understood. While...
5
24
108
Enjoyed presenting at #NGSB24! Thanks to the entire OC for putting together a fantastic meeting! So many exciting new avenues for SynBio, from DNA synthesis, cell-free systems, designing regulatory systems, all the way to improving AI and exploring new applications!.
Afternoon session now at #ngsb24 - and it’s the first of four talks on Machine Learning and AI in synbio. Really nice first talk by Judith Kribelbauer from EPFL
1
0
11
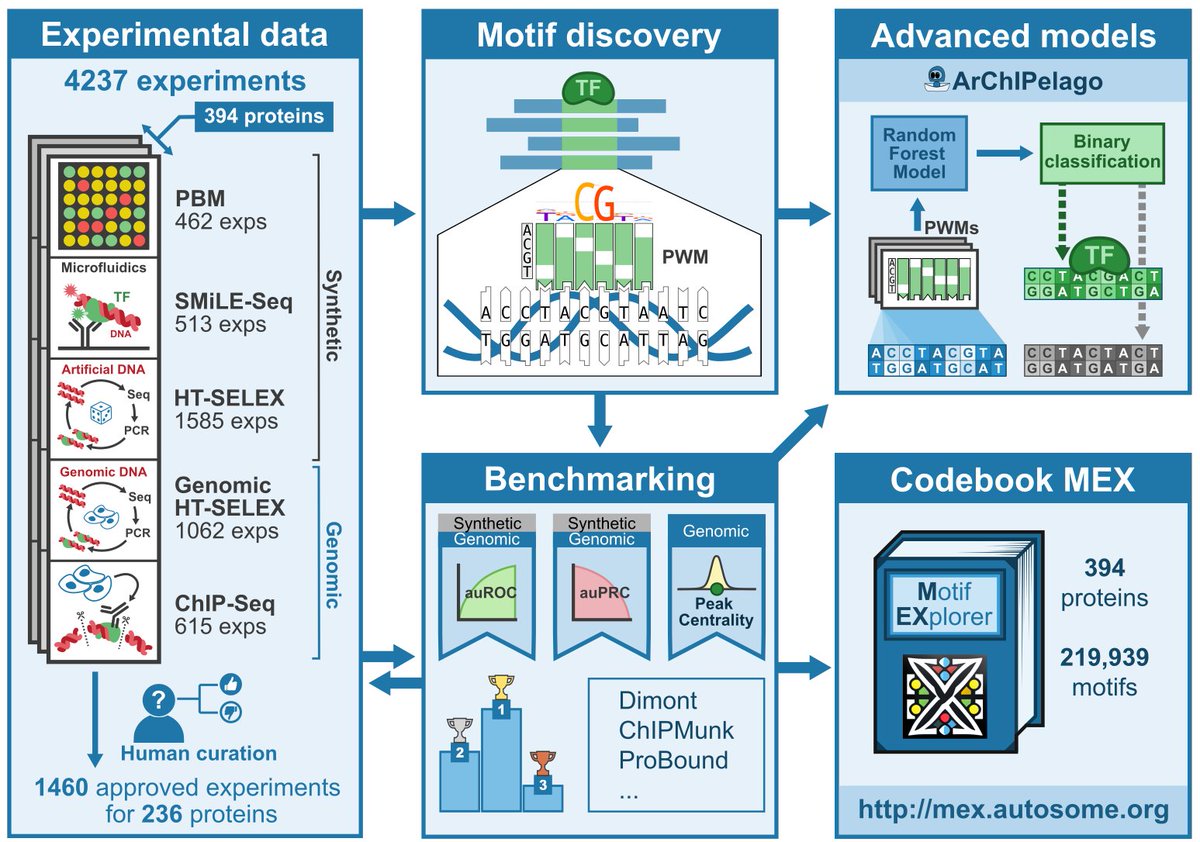
RT @ArttuJolma: (1/8) Thrilled to reveal the results of the Codebook Project, an international effort to identify accurate DNA-binding moti….
0
8
0
Check out this preprint from @AntoniGralak a very talented PhD in the @BartDeplancke lab, developing methyl-SMiLE-seq and applying it to study poorly characterized human TFs as part of the Codebook Consortium! 1/5 linked preprints across exp. platforms.
0
0
4
It's finally out! Massive team effort by the Codebook Consortium to characterize binding specificities of poorly studied human TFs. Preprint below highlights results of the benchmarking (1/5 preprints). Glad I was able to contribute as part of @BartDeplancke lab, w @AntoniGralak.
(1/13) Excited to present the results of the large-scale benchmarking of DNA motif discovery tools using the Codebook data compendium on poorly studied human transcription factors and the Codebook Motif Explorer .| ️
0
0
16
highly recommend!.
New synthetic biology / gene regulation lab opening in Zurich! We’re studying how to control cell state transitions – for example, making diseased cells healthy or creating new cellular functions – using systematic perturbations, single-cell genomics and machine learning. 🧵1/4
1
0
2
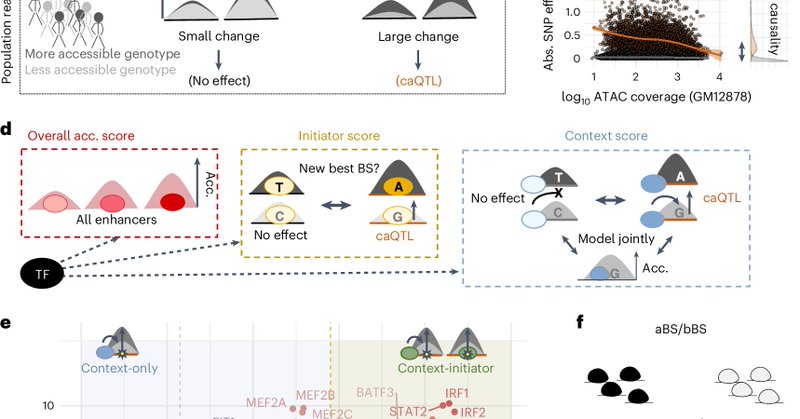
Excited to have our work out now in @NatureGenetics. @BartDeplancke . Few important changes compared to the preprint! see 🧵 below.
nature.com
Nature Genetics - This study identifies context-only transcription factors (TFs), a TF class that enhances DNA accessibility initiated by cell type-specific TFs and establishes cooperative...
9
26
124
Excited to share our latest tool at #NGSB24, leveraging synthetic enhancers to learn what deep learning models get right & wrong about how the non-coding genome controls gene expression. Pre-print coming soon!.
🧬Are you a researcher working in #SyntheticBiology? Then, present your latest breakthroughs to key scientists in this exciting field at #NGSB24. Plenary poster pitches & the @EMBO Poster Prize are up for grabs!.⬆️Upload your abstract before 8 Oct: #Synbio
0
1
7