Arttu Jolma
@ArttuJolma
Followers
55
Following
37
Media
5
Statuses
25
Research Associate in lab. of Tim Hughes. Studying transcription factor target specificities, gene regulation and RNA binding proteins.
Toronto, ON, Canada
Joined January 2018
(8/8) . @IvanDKoz, @nikitagryzunov, @VorontsovIE, @halfacrocodile, @legoushque, @AntoniGralak, @ZinkevichA, @JKribelbauer and see also
0
0
0
(7/8) Kudos to the Codebook and GRECO-BIT consortium members including @BartDeplancke, @MakeevSeva, @EPD_SIB, @OFornes, @ImYellan, . .
1
0
0
I was going to make a Tweetorial describing our new paper, but it looks like Hamed covered it already very well. Enjoy!.
Super excited to finally share the results of a massive effort with @ProfTimHughes to characterize the intrinsic sequence binding specificities of human transcription factors, led by @ArttuJolma, @ahcorcha, @AliFathi125, Ally Yang, and Kaitlin Laverty 1/n.
0
1
8
Nice to see my new work getting some publicity, I'll try to get a Tweetorial done for this in the near future.
GHT-SELEX demonstrates unexpectedly high intrinsic sequence specificity and complex DNA binding of many human transcription factors (Jolma et al., 2024) .▶️TFs possess sufficient intrinsic specificity to independently delineate cellular targets
0
1
13
RT @generegulation: Extensive binding of uncharacterized human transcription factors to genomic dark matter (Razavi et al., 2024) https://t….
0
54
0
So here is a brand new study with my second author contribution. It is a cool story about how most of the RNA binding observed in mass spec studies for a large fraction of all proteins is in fact is nonspecific.
✨New paper alert✨. We assayed the binding specificities of 492 unconventional RBPs (ucRBPs) with RNAcompete and found that most bind non-specifically.
0
1
9
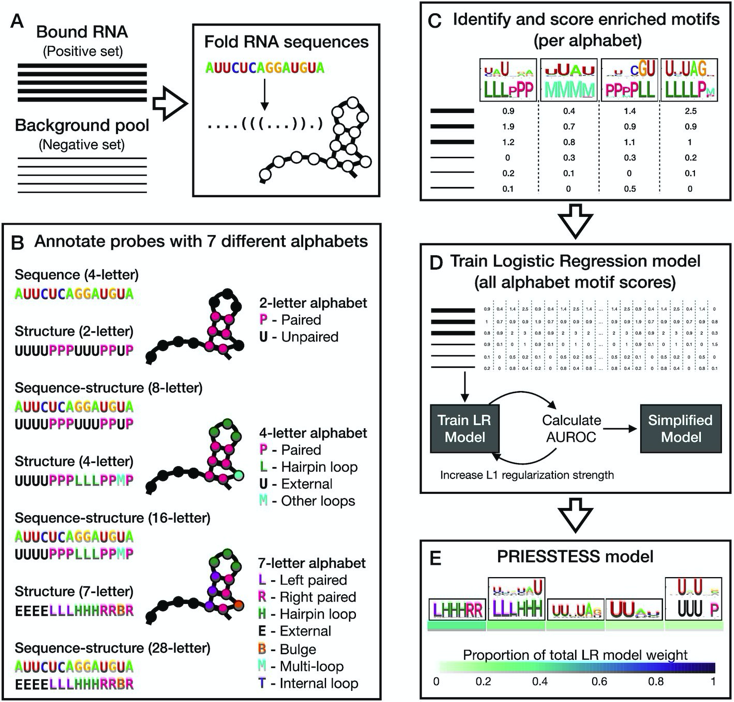
RT @quaidmorris: Check out our new RBP motif algorithm: .PRIESSTESS 🧙♀️.- fits RNA sequence and structure models .- is trivially interpret….
0
37
0
Yarr, congratulations to my boss!.
Congratulations to all @UofT faculty who won Canada Research Chairs but above all to our amazing Prof Tim Hughes!. The goal of his research is to be able to read the genome the way cells do. Find out more:. @CRC_CRC @uoftmedicine
0
0
1
My ex-boss Jussi Taipale and and another Swedish Professor Sten Linnarson are proposing a very sensible strategy for combatting Covid-19. We should make testing a massively parallel high throughput process and perform it repeatedly on the scale of whole population.
”To stop COVID-19, test everyone, repeatedly.” Me and Jussi Taipale propose a simple strategy to defeat coronavirus, and pose a challenge to the scientific community.
0
0
0
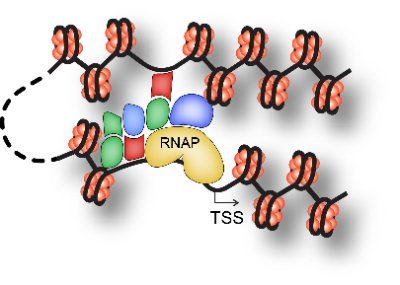
RT @Gene_Regulation: Strong binding activity of few transcription factors is a major determinant of open chromatin .
0
133
0