Jeroen Gilis
@GilisJeroen
Followers
220
Following
1K
Media
13
Statuses
368
PhD student, developing statistical software for single-cell transcriptomics analysis, Ghent university
Belgium
Joined June 2019
I am now a Doctor of Philosophy in Statistical Data Analysis! 🎉🥳 I am very grateful to have had three wonderful supervisors (@lievenClement , @YvanSaeys and @koenvdberge_Be ) by my side. The result is several publications ( https://t.co/pO0k8FDOVG), and finally, an PhD thesis.
2
0
14
Strategies for addressing pseudoreplication in multi-patient scRNA-seq data https://t.co/Wyi7Wv5zvk
#biorxiv_bioinfo
0
2
4
I also want to thank the members of my PhD jury (@drisso1893 , Charlotte Soneson, @tidmeyer, @kdpreter, @SVansteelandt and Jan De Neve) for their insightful remarks and comments on the thesis 🙏
0
0
1
📢New preprint from our lab Self-supervised learning for fluxomic data preserves structure across cell types @cjmerzbacher @ancAtEd
#sysbio #synbio #ML #metabolicmodelling #cobrapy
https://t.co/9Nun3a7XSJ
0
7
25
I have several exciting openings for PhD students and postdocs in the context of my advanced ERC grant #ERCAdG on Assumption-lean (Causal) Modeling and Estimation. Please spread the news! https://t.co/zLP5Z93FyN
@DassGhent
@UGent_fwe
@ResearchUGent
@UGent
@ERC_Research
0
23
38
After four years of user feedback and experience, our NicheNet best practices guideline is now available on arXiv! https://t.co/GsVdxO3fdR Also check out our revamped GitHub repository that incorporates these changes: https://t.co/Dc9odQ4ini (1/2)
1
12
47
Exhilarated to announce I've been tenured at @DTUtweet 🤯🥳. Thanks to @GreeneScientist and @tuuliel for taking the time to evaluate my application 🙏 The timing is perfect, as I have 3 bioinformatic openings (both PhD and postdoc) - see👇 Retweets are sincerely appreciated 😊
7
8
37
Papers submissions open for “Advancements in Biological Network Analysis”, at CIBB 2024 (19th conference on Computational Intelligence Methods for Bioinformatics and Biostatistics), September 4-6, Benevento, Italy. Deadline: 15th May https://t.co/2BHsYsxSNx
0
2
3
📢new preprint from our lab: Multiscale pathway simulation coupled with host metabolism. It combines genome-scale, kinetic and #MachineLearning models. https://t.co/lKyfU1PO0n Fab work by @cjmerzbacher with @oisinmacaodha
#syntheticbiology #AI #Bioinformatics
0
14
43
I am deeply honored and humbled to be recipient of an advanced ERC award #ERCAdG to spearhead research on Assumption-lean (Causal) Modeling and Estimation. @DassGhent
@UGent_fwe
@ResearchUGent
@UGent
@ERC_Research 1/2
9
6
70
🧬Easily & effortlessly generate communities of genome-scale metabolic models with PyCoMo for #COBRA ‼️Plus: Fully compliant w #SBML standards‼️ Check out https://t.co/itTDYcJXym Excellent collaboration lead by #MichaelPredl & #MarianneMiesskes w #CUBE @univie_stem ⚙️
0
7
13
How can we improve trustworthiness of cell type classifiers ? Well, for example by combining uncertainty quantification with hierarchical models with a reject option. Read all about it in Lauren Theunissen's first paper at https://t.co/kerH9tBv2j
#AIFlanders
1
10
40
Excited to talk about our long-read benchmarking work next week!
Next Monday, @DongXueyi from @WEHI_research will present: Benchmarking long-read RNA-sequencing analysis tools using in silico mixtures ⏱️March 25, 1pm (AEST) 👉 https://t.co/wvVjkPCBZ6
0
4
25
If you’re interested in learning more about single-cell and spatial transcriptomics and proteomics, join us for CSAMA 2024 in the beautiful Italian Alps! List of faculty, topics, and registration info at
csama2024.bioconductor.eu
Biological Data Science, 23-28 June 2024, Brixen / Bressanone, Italy
0
9
14
Alternative splicing as a source of phenotypic diversity https://t.co/fZbWAtEQK3
#Review by @CharJWright, Christopher W. J. Smith & @mel_rosina
@sangerinstitute @darwintreelife @CamZoology @CamBiochem @LabsRna
nature.com
Nature Reviews Genetics - In this Perspective, the authors discuss how regulated alternative splicing can generate phenotypic diversity and outline emerging evidence that alternative splicing...
2
72
258
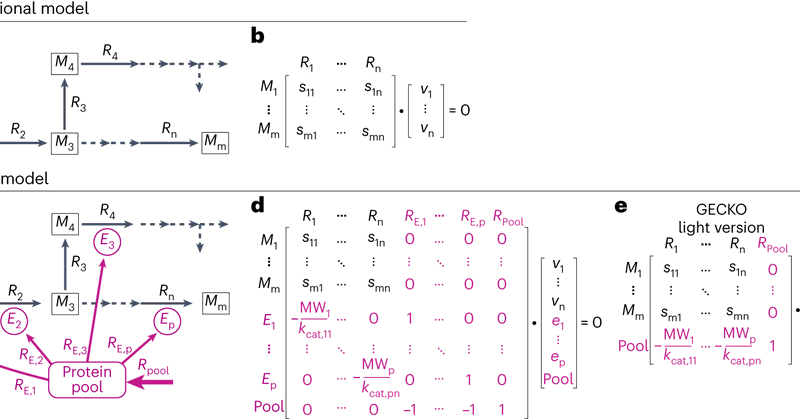
#FeaturedProtocol: Reconstruction, simulation and analysis of enzyme-constrained #metabolicmodels using GECKO toolbox 3.0 https://t.co/ZGSfAZuH4s
#iclip2
nature.com
Nature Protocols - Genome-scale metabolic models enable mathematical exploration of metabolism under various defined conditions. This protocol describes GECKO, a method for enhancing a genome-scale...
0
4
5
.@hector_rdb, @koenvdberge_be, @justokstreet & @cendrinou present condiments for the inference and downstream interpretation of cell trajectories across multiple conditions #BiotechNatureComms
https://t.co/ovnoqy5yEz
nature.com
Nature Communications - scRNA-Seq has enabled the study of dynamic systems such as response to a drug at the individual cell and gene levels. Here the authors introduce a framework to interpret...
0
3
9
Why do Random Forests perform so well off-the-shelf & appear essentially immune to overfitting?!? I’ve found the text-book answer “it’s just variance reduction 🤷🏼♀️” to be a bit too unspecific, so in our new pre-print https://t.co/UXDO9ULnl6,
@Jeffaresalan & I investigate..🕵🏼♀️ 1/n
13
213
1K
Interested to know how #reprocessing and reusing existing #proteomics #data helps uncover the unknown? Let's have a beer and talk about this at @DataBeersBru this Thursday. Details below! See you soon! #bigdata #humanproteome
✨Our 3rd #databeers #brussels speaker will be @Pathmanaban_ with "Unveiling Protein Mysteries through Advanced Data Mining and Re-processing" Our waiting list is open ➡️ https://t.co/ClahHBDXd5
0
4
8
Our website https://t.co/Ql2OzyQ9BT had an upgrade thanks to @BaryonDesign. Read now our latest blog post about our microplastic research in agricultural fields! #microplasticresearch #erc
0
2
2