Georg Krainer
@GeorgKrainer
Followers
508
Following
422
Media
6
Statuses
147
Research Group Leader in Molecular Biophysics @UniGraz | Senior Scientist & Founding Team Member #TransitionBio
University of Graz, Austria
Joined October 2018
👀 Exciting times ahead. Stay tuned for new discoveries and hiring opportunities.
0
0
1
My heartfelt thanks go to @UniGraz, along with my wonderful mentors, collaborators, colleagues, friends, and family for their unwavering support. https://t.co/YXUQEuXHfd (German)
uni-graz.at
Die Universität Graz sichert sich drei ERC Starting Grants. Diese Auszeichnungen bestätigen die starke Position der Universität im Forschungswettbewerb.
1
0
1
Using cutting-edge single-molecule and microfluidic techniques alongside theoretical modeling, we aim to offer a fundamentally new biophysical perspective on GPCR signaling, ultimately opening new avenues for drug design guided by physical principles.
1
0
1
A huge thank you to @ERC_Research for supporting my mission to uncover how G protein-coupled receptors (GPCRs) function at the molecular level.
1
0
1
🚀 Exciting news! 🚀 I’m thrilled to share that I’ve been awarded an ERC Starting Grant for my project SignAlloMod: Decoding the Molecular Logic of GPCR Signaling! https://t.co/ZgjSL8DsEp
erc.europa.eu
The European Research Council (ERC) has announced the award of 494 Starting Grants to young scientists and scholars across Europe. The funding - totalling nearly €780 million - supports cutting-edge...
8
0
34
Big thanks to all co-authors, especially @BJacquat @SchneiderMatth @TimWelsh_ #JennyFan @KnowlesLabCamb & collaborators @SteveTheChemist @Alberti_Lab #HartlLab #VasilisKosmoliaptsis
0
0
3
📃Excited to share our preprint on single-molecule digital sizing of proteins. 🔥Highlights: sizing at fM concentrations, affinity profiling at the single-molecule level & sizing of polydisperse sample mixtures (oligomers, aggregates, nanoclusters)🚀🧪⤵️
biorxiv.org
Proteins constitute the molecular machinery of life and exert their biological function by interacting with other proteins, as well as by assembling into biomolecular complexes and higher order...
2
5
37
Work led by amazing PhD student @HAusserwoeger @KnowlesLabCamb @ChemCambridge in collaboration with @novonordisk and @CCG_MOE . Such a fun and intriguing project with new discoveries evolving until the very end, and more to follow. Stay tuned!
0
2
2
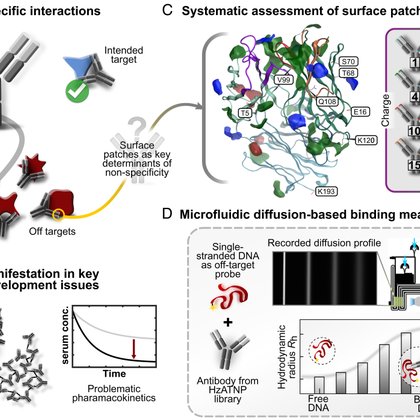
👇📰Check out our new paper "Surface patches induce nonspecific binding and phase separation of antibodies". 😎We looked at nonspecific binding of antibodies to DNA 🧬, and found that sticky surface patches can lead to antibody phase separation💦.
pnas.org
Nonspecific interactions are a key challenge in the successful development of therapeutic antibodies. The tendency for nonspecific binding of antib...
Check out our new work in @PNASNews on how surface patches control protein binding specificity: https://t.co/JdOHnQRp1N Great work by @HAusserwoeger, @GeorgKrainer, @TimWelsh_, @ElladeCsillery, Tomas, @SchneiderMatth and Therese in collaboration with @novonordisk and @CCG_MOE.
1
1
9
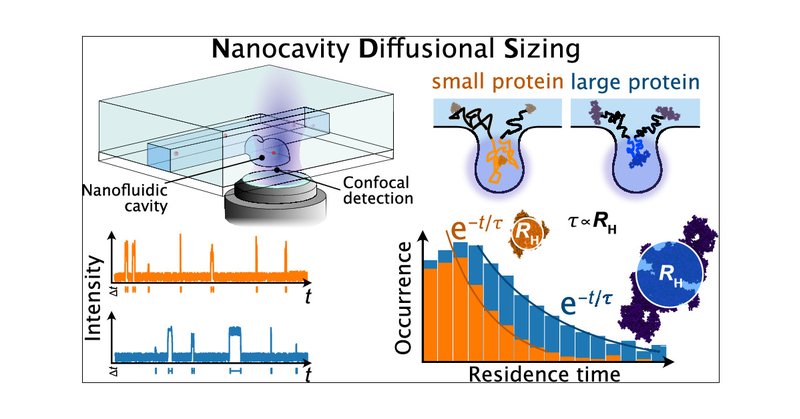
🔓 #OpenAccess + Most Read in the past 30 days: "Single-Molecule Sizing through Nanocavity Confinement" by @BJacquat, @GeorgKrainer, Peter, Babar, Vanderpoorten, Xu, Welsh, Kaminski, Keyser, Baumberg, & Knowles https://t.co/idonqg5HSq
pubs.acs.org
An approach relying on nanocavity confinement is developed in this paper for the sizing of nanoscale particles and single biomolecules in solution. The approach, termed nanocavity diffusional sizing...
0
5
12
Bored of laborious and time consuming single-well #smFRET measurements? Check out our recent preprint on multiwell plate #smFRET 👇 Great work led by @HartmannPHY @Schlierf_Lab
biorxiv.org
Single-molecule FRET (smFRET) has become a widely used tool for probing the structure, dynamics, and functional mechanisms of biomolecular systems, and is extensively used to address questions...
Farewell to single-well #smFRET. Our newest story on bioRxiv https://t.co/XwvHNXeMWG Great team work led by @HartmannPHY with @KoushikS33, Mathias, @nehagc18, Philipp and @GeorgKrainer. Brief tweetorial and open access software 1/5
0
0
13
📃 Our latest work is out! Sizing of single proteins using nanofluidics. 🧪 Great collaboration with @BJacquat and many others @KnowlesLabCamb @ChemCambridge & in collaboration with #baumberglab,@KeyserLab & @group_laser
pubs.acs.org
An approach relying on nanocavity confinement is developed in this paper for the sizing of nanoscale particles and single biomolecules in solution. The approach, termed nanocavity diffusional sizing...
0
11
42
@KnowlesLabCamb @ChemCambridge Big props to @KadiLiisSaar, twitterless #WillArter, @TimWelsh_, @magdaczek & many more including @KlenermanLab, @biochemist_hero & twitterless #AlbertiLab
0
1
4
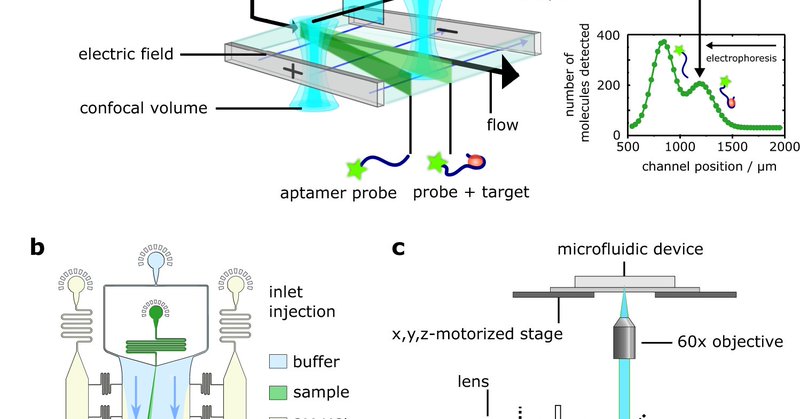
📢 Finally out now: Our paper describing DigitISA, a platform for direct digital sensing of protein biomarkers in solution 🧪 (incl. antibodies, amyloid aggregates & condensates). @KnowlesLabCamb @ChemCambridge Take a look: 👇 https://t.co/j6MycHGG5t
nature.com
Nature Communications - There are limitations with current protein sensing methods. Here the authors report DigitISA, a digital immunosensor assay based on microchip electrophoretic separation and...
1
3
18
What a month. Two starting grants submitted 📃 , and now been promoted to father. 👼 ;) #helloworld #bundleofjoy #dreamsdocometrue ❤
4
0
38
So incredibly thrilled and honored to have received the Young Investigator Award by the German Biophysical Society! Thank you #DGfB for this high recognition of my work! And special thanks to my mentors @KnowlesLabCamb @Schlierf_Lab and twitterless #SandroKeller 🥳 #biophys2022
6
2
48
Check out our preprint discovering DNA-induced LLPS of antibodies based on surface patch driven non-specific interactions: https://t.co/VrHFHnYOun Great work by @HAusserwoeger, @GeorgKrainer, @TimWelsh_, Tomas, @SchneiderMatth & Therese. Exciting collaboration with @novonordisk
biorxiv.org
Non-specificity is a key challenge in the successful development of therapeutic antibodies. The tendency for non-specific binding in antibodies is often difficult to reduce via judicious design and,...
0
4
24
..and a preprint on Multiphase architectures in single-component condensates 💦: https://t.co/SP5e6VXt1X with @rcollepardo #lab @KnowlesLabCamb @ChemCambridge
0
0
2
LLPS in rotavirus replication factories 🦠: https://t.co/zKUWlWzLSz with @AlexBoRNA #lab An all-aqueous LLPS system bioengineered from protein microgels 🪄: https://t.co/mS4ZdW9AGb
@KnowlesLabCamb @ChemCambridge
embopress.org
image image Early replication factories provide a microenvironment for replication of RNA viruses. Here, rotavirus replication factories are found to represent protein/RNA‐rich condensates formed by...
1
0
3
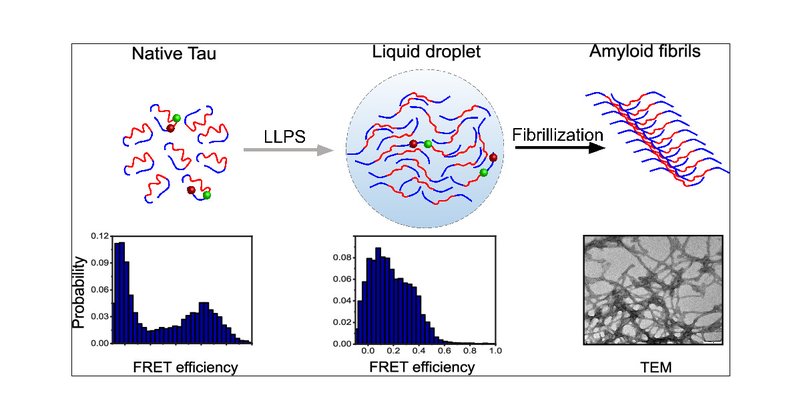
Been busy lately 🧑🌾, totally failed to mention a few recent 📰papers: α-synuclein fibril disaggregation by Hsc70 🤖: https://t.co/Ao8ZGMJh4y with #HartlLab Conformational expansion of Tau during LLPS 🦄: https://t.co/CgcGOUEzKL with #PerrettLab
@KnowlesLabCamb @ChemCambridge
pubs.acs.org
Liquid–liquid phase separation (LLPS) of proteins into biomolecular condensates has emerged as a fundamental principle underpinning cellular function and malfunction. Indeed, many human pathologies,...
1
2
24