Frydman Lab
@FrydmanLab
Followers
1K
Following
160
Media
16
Statuses
196
A lab at Stanford University studying protein biogenesis, folding, and aggregation in health and disease. Tweets by lab members.
Stanford, CA
Joined February 2020
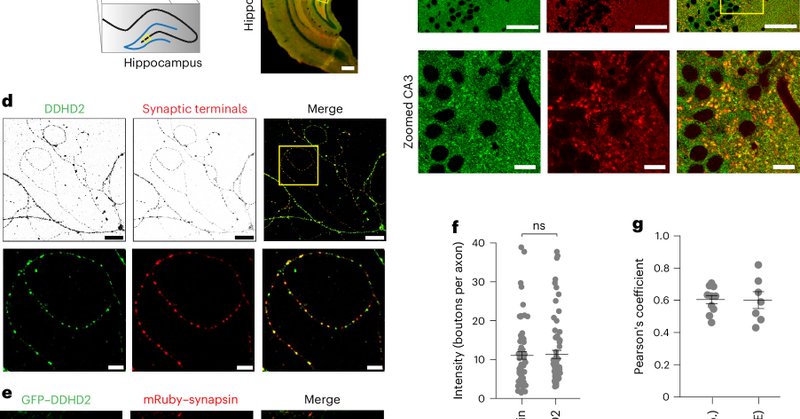
Another great story from Jae Ho Lee in the lab: a new concept in cotranslational proteostasis-ribosome communication via chaperoneNAC. An exciting collaboration with Elke Deuerling's lab @DeuerlingLab and Marina Rodnina's lab
biorxiv.org
The nascent polypeptide-associated complex (NAC) is a conserved ribosome-bound factor with essential yet incompletely understood roles in protein biogenesis. Here, we show that NAC is a multifaceted...
1
11
30
New exciting work from Jae Ho Lee in our lab, in a fantastic collaboration with Alessandro Ori @AOri_lab and Alessandro Cellerino @Alessan82703458 on why and how the brain ages and becomes vulnerable to neurodegenerative diseases
science.org
Aging is a major risk factor for neurodegeneration and is characterized by diverse cellular and molecular hallmarks. To understand the origin of these hallmarks, we studied the effects of aging on...
0
15
49
RT @anshulkundaje: Paging @NIHDirector_Jay . Is abruptly cutting these grants going to "make America healthy again"?.
0
5
0
RT @NatMetabolism: Triglycerides are an important fuel reserve for synapse function in the brain
nature.com
Nature Metabolism - Kumar et al. show that under glucose-depleted conditions, neurons can use fatty acids as an alternative source of energy to support synaptic function.
0
61
0
RT @JMaraganore: I stand with dozens of other biopharma leaders highlighting the potential catastrophic effects of proposed research cuts o….
statnews.com
Proposed cuts will have “a catastrophic effect on the advancement of biomedical and biotechnology capabilities in the United States,” more than 100 leaders of biomedical/health sciences companies...
0
4
0
RT @JohnStreicher1: My NIDA grant renewal is almost 2 months late and I’m going to have to start firing my students and staff. Not a lot of….
0
57
0
RT @segal_eran: Our lab at the Weizmann Institute was hit tonight by Iran. We will rebuild and return 💪
0
647
0
RT @PiereBiophysics: It was a pleasure to participate in the "Protein folding & Chaperones" platform session this morning in #bps2025 shari….
0
1
0
RT @FaboPolanco: Closing an unforgettable chapter at Stanford. Grateful for the science, the growth, and the incredible people who shaped t….
0
3
0
Check out our MS describing the in vivo cycle of chaperonin TRiC/CCT, in a wonderful collaboration with Martin Beck's lab: 1) The study established TRiC's important function in folding newly translated proteins and defines its ATP-driven cycle.
nature.com
Nature - TRiC functions at near full occupancy to fold newly synthesized proteins inside cells.
0
12
48
4. The concept of chaperonin function as central in brain development is a new paradigm opening the door to many additional mysteries. Thanks to all: Ingo, Miriam, Stephen, Florian, Piere for the fun collaboration and to Manu Sharma for the highlight:
science.org
Mutations that impair a protein-folding chaperone can lead to brain malformations
0
6
31
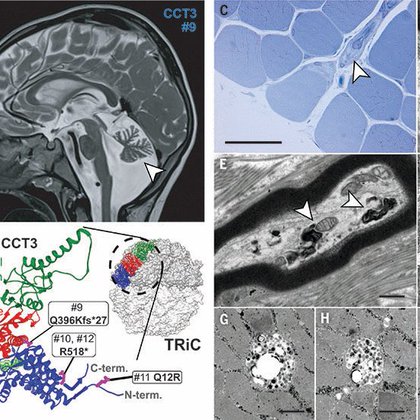
1. Check out our latest and exciting MS describing a new class of neurodevelopmental diseases caused by mutations in most subunits of the chaperonin TRiC/CCT. (part 1 of series).
science.org
Malformations of the brain are common and vary in severity, from negligible to potentially fatal. Their causes have not been fully elucidated. Here, we report pathogenic variants in the core protei...
2
16
69