Alex Dunn
@Dunn_Lab
Followers
1K
Following
94
Media
0
Statuses
94
Joined December 2018
New preprint! @vvachhar and @DejongMatt show that PDZ domains, innocuous-looking scaffolding domains that are enriched at cell-cell junctions, can exhibit striking forms of mechanosensitivity:. (1/2).
biorxiv.org
Intercellular adhesion complexes must withstand mechanical forces to maintain tissue cohesion, while also retaining the capacity for dynamic remodeling during tissue morphogenesis and repair. Most...
4
7
41
New paper! @clhueschen presents a mathematical model of flocking on arbitrary 2D surfaces. In addition to your favorite creature, the model can be applied to a broad range of problems in active matter physics, e.g. cytoskeletal flows in living cells.
journals.aps.org
The collective motion of animal herds and other biological systems sometimes takes place in an environment with a complex geometrical structure. In this paper, the authors study such active matter...
0
7
42
RT @Leeya_Engel: We micropatterned lymphocytes on EM grids to facilitate cryo-FIB & cryo-ET, revealing networks of filaments on the Jurkat….
0
13
0
New preprint: @bri_zhong shows that pN forces can potentially activate latrophilin 3, an adhesion GPCR that regulates synaptic plasticity. A first step for the lab in understanding this fascinating class of signaling proteins. Congrats Brian!.
biorxiv.org
Latrophilins are adhesion G-protein coupled receptors (aGPCRs) that control excitatory synapse formation. aGPCRs, including latrophilins, are autoproteolytically cleaved at their GPCR-Autoproteolysis...
1
8
50
RT @AnneStraube: #MotorsInQuarantine will feature @amywang01 from @Dunn_Lab and @MarkDodding. Join us tomorrow to discuss how actin links t….
0
9
0
New paper! With the @HTSohLab, we show how single-molecule imaging can be used to to suppress background signal in sandwich-type immunoassays. Congrats and thanks to @AmaniHariri20, @SharonNewman0, and @steventan17 for a great project and collaboration.
0
0
16
Just out: MBoC special issue on mechanobiology! Many thanks to @MattWelchLab and co-editors Dennis Discher, Valerie Weaver, @TheYapLab, and @murrell_lab for involving me in this project.
molbiolcell.org
0
1
25
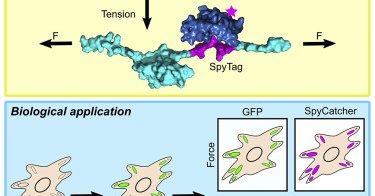
New paper! @bri_zhong describes a new strategy for visualizing mechanical forces across proteins in cells. We hope this will be a useful tool for everyone interested in mechanobiology and are excited to build upon this work in the future.
cell.com
Zhong et al. describe STReTCh, a genetically encoded force-sensing module that provides a readout of molecular forces in biological systems. STReTCh relies on typical “fix-and-stain” immunofluoresc...
1
37
177
RT @Henry_de_belly: Excited to share that @cavankatenaugh. (@OWeinerLab ), Jennifer Hill (@DavidGDrubin ) and I are organizing the 2022 e….
0
9
0
Thanks to some good funding news, the lab is looking for postdocs for projects with @LabSkiniotis and Bill Weis (Stanford Struct. Bio.). We'll be exploring ideas I'm quite excited about at the interface of biophysics, cell, and structural biology. Please email for more details!.
0
14
43
New paper! @amywang01 describes the probable mechanism by which alpha-catenin, and likely related proteins, makes a force-sensitive catch bond with F-actin. Also, intriguing clues about the origins of cooperative actin binding. Congrats Amy!.
elifesciences.org
Deleting the first helix of the α-catenin actin-binding domain results in a slip bond interaction between the cadherin–catenin complex and F-actin, such that the binding interaction is stable at low...
3
18
100
The mathematical tools Cayla and @ElginKorkmazhan developed should be applicable to any situation where noisy single-molecule data are being fit to an underlying statistical distribution. Congrats, Cayla and Elgin! (2/2).
0
0
7
New paper! @CaylaMMiller used a clever approach to extract the distribution of actin filament velocities in living cells. The data support a model of the actin cytoskeleton in which periods of mechanical equilibrium are interrupted by sudden jumps. (1/2).
1
17
100
New paper! .This surprising finding suggests one way by which the cell generates stable linkages between the membrane and cytoskeleton while still allowing rapid cytoskeletal remodeling. Congrats @ElginKorkmazhan!.
2
27
131
RT @cavankatenaugh: PhDs and Postdocs: we are still accepting abstracts for the GRS on Signaling from Adhesion Receptors - we will be exten….
0
35
0
Tweetorial by Elgin on his recent paper that I tweeted about a few days ago. Please enjoy the awesome movies!.
HOW do cells respond to being peeled off e.g. when you pinch yourself too hard? We find that once cells partly detach, newly extruded membrane tethers attached to the surface (e.g. retraction fibers) guide cells back to their original location 1/
0
1
22
In collab. with Andrew Kennard (@askennard), Elgin shows that a similar process occurs in a zebrafish wounding model. (3/4).
1
0
4