Deen Sun
@DeenSun3
Followers
420
Following
6K
Media
1
Statuses
526
Incoming assistant professor | Postdoc in Kai Johnsson Lab @mpi_mr_hd | Ph.D in @XingChen_PKU Lab @PKU1898 | Chemical biology, Expansion🔬, Molecular labeling
Heidelberg, Germany
Joined January 2018
Our biorxiv preprint is now online @nchembio 🎉 https://t.co/PIY7kk2APJ We made substantial revisions and added new experiments. Thanks to everyone who made this possible. Our journey continues!
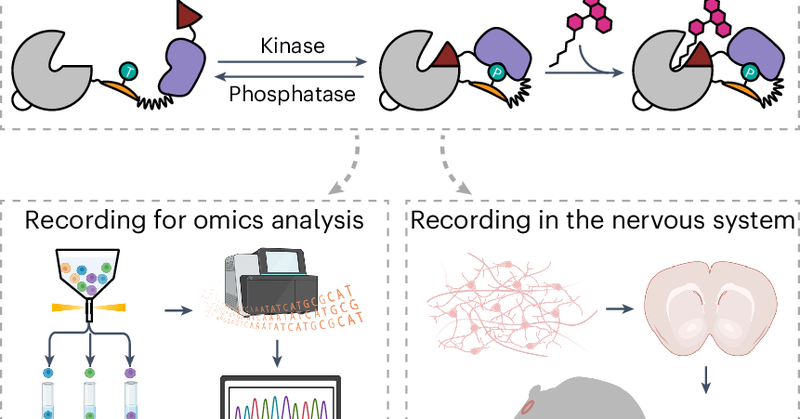
🚨Excited to share my latest postdoc work in this #biorxiv preprint on developing split-HaloTag recorders for capturing cellular transient kinase activity, Kinprola (kinase activity dependent protein labeling). https://t.co/iAUx1XG8gj 🧵 (1/7)
2
3
30
🚨Excited to share this collaborative work with Ming-Ming. Check it out!🎉▶️ https://t.co/YFlhCncvHs
biorxiv.org
Elucidating the role of metal ion homeostasis in physiological and pathological processes requires detection tools with high sensitivity and selectivity, spectral versatility, and precise subcellular...
New paper out! We developed localizable chemigenetic fluorescent sensors for key metal ions (K⁺, Na⁺, Ca²⁺, Mg²⁺, Zn²⁺, etc.). Our K⁺ indicators work beautifully in live cells and neurons. Samples available on request. Huge thanks to @DeenSun3 and Kai @mpi_mr_hd! Link 👇
0
0
3
Our November issue is live! https://t.co/dU6JE66vS0 The cover depicts a SEM image of a dorsal surface of liverwort M. polymorpha female thallus. Liverwort were found to have a noncanonical pathway for the regulation of master growth repressor DELLA.
0
9
21
Our RAEFISH spatial transcriptomics technology is now published in Cell @CellCellPress! RAEFISH enables sequencing-free whole genome spatial transcriptomics at single molecule resolution. This work represents the first time that transcripts from more than 23,000 genes were
17
118
412
Molecular recording of cellular protein kinase activity with chemical labeling:
nature.com
Nature Chemical Biology - Molecular recorders based on kinase activity-dependent protein labeling track specific kinase activities to understand their link to cellular phenotypes in heterogeneous...
0
51
184
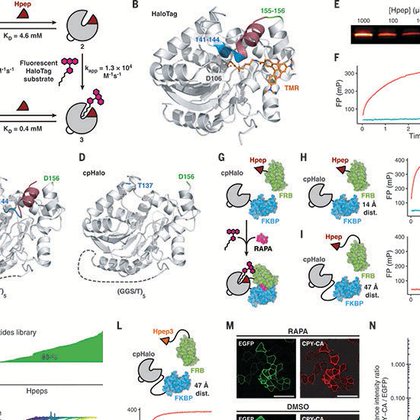
🚨 New preprint alert! Looking for a peptide tag alternative to the classic HaloTag for protein labeling? Check out our latest work "A high-affinity split-HaloTag for live-cell protein labeling" on #bioRxiv 🔗 https://t.co/P5H3DzHITf
biorxiv.org
We introduce a high-affinity split-HaloTag comprised of a short peptide tag (Hpep, 14 residues) and a large, inactive fragment (cpHaloΔ3). Hpep binds to cpHaloΔ3 spontaneously with nanomolar affini...
2
6
18
Looking for a fluorescent biosensor and don’t know where to start? Want a comprehensive overview of the field? Check out our new review in @ACSChemRev online now as an ASAP article! From co-first authors @AMMGest @AyseZSahan @yanghao_zhong
https://t.co/YId0ttkqkf
2
30
99
Thanks especially to Kai Johnsson and to all involved! On X you will find @DirkCHoffmann @KimBoonekamp @Winkler_Lab @wick_wolfgang @Michael_Boutros @yulonglilab @mpi_mr_hd @EPFL_en @uniklinik_hd @UniHeidelberg @PKU1898 @AvHStiftung @ResearchLifesci (7/7)
0
0
1
Kinprola enables the recording of PKA activation in primary neurons, acute brain slices upon drug or electrical stimulation, and for the tracking of drug-induced neuromodulation in freely moving mice🐭. (6/7)
1
0
1
Kinprola enables the recording of differential PKA activities in heterogeneous cell cultures after pooled genetic perturbation🧬✂️, for genome-wide CRISPR screens to discover regulators of PKA signaling. (5/7)
1
0
0
We applied Kinprola for capturing differential PKA activities within the heterogeneous network of glioblastoma cells, enabling to separate cells by FACS based on their kinase activity for RNA-Seq analysis. (4/7)
1
0
0
The fluorescent mark can be chemically fixed and analyzed using flow cytometry or fluorescence microscopy🔬. We have designed specific recorders for four protein kinases, including protein kinase A. (3/7)
1
0
0
Based on the split-HaloTag system we developed earlier this year ( https://t.co/rnH3ZIJkGN) and a phosphorylation-dependent molecular switch, Kinprola become rapidly labeled in the presence of a specific kinase activity and a fluorescent HaloTag substrate. (2/7)
science.org
Modified proteins allow recording of a cell’s regulatory activity at a specific period of time for later analysis.
1
0
0
🚨Excited to share my latest postdoc work in this #biorxiv preprint on developing split-HaloTag recorders for capturing cellular transient kinase activity, Kinprola (kinase activity dependent protein labeling). https://t.co/iAUx1XG8gj 🧵 (1/7)
biorxiv.org
Protein kinases control most cellular processes and aberrant kinase activity is involved in numerous diseases. To investigate the link between specific kinase activities and cellular phenotypes in...
2
13
89
An inspiring read! Synthesis at the Interface of Chemistry and Biology | Accounts of Chemical Research
pubs.acs.org
ConspectusChemical synthesis as a tool to control the structure and properties of matter is at the heart of chemistry─from the synthesis of fine chemicals and polymers to drugs and solid-state...
2
10
39
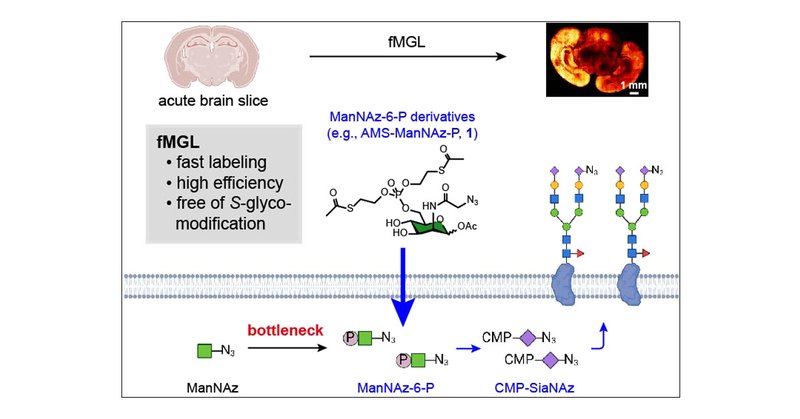
Glad to contribute to this work led by my phd lab @XingChen_PKU Fast Metabolic Glycan Labeling for Click-Labeling and Imaging of Sialoglycans in Living Acute Brain Slices from Mice and Human Patients | Journal of the American Chemical Society
pubs.acs.org
Living acute brain slices provide a practical platform for imaging sialylation in human brain pathology. However, the limited lifespan of acute brain slices has impeded the use of metabolic glycan...
0
0
3
🔬Join us at #ChemoRevolution, where chemistry meets biology to reshape our understanding of life! Explore cutting-edge tools and groundbreaking discoveries in chemical biology. Connect with global experts and rising stars in the field. Be part of the scientific revolution!
0
4
21
a very nice outlook
Proximitomics by Reactive Species | ACS Central Science https://t.co/Q1O9W588qO Our recent Outlook is out now. We reviewed recent advances in proximity labeling at multiple scales with reactive species. Check it!
1
0
3
Our new #MPIMRSpotlight features @DeenSun3, postdoctoral researcher in the Chemical Biology department! ⚗️Check out what he has to say about his work at the Institute and interest in spying on brains!🔎🧠 👀
0
2
17
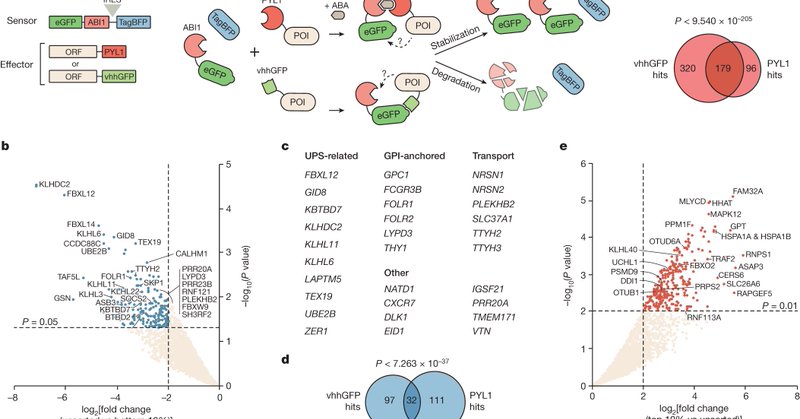
Our paper on functional discovery of protein degradation and stabilization effectors with proteome-scale induced proximity screens is out in its final version! 1/ https://t.co/vzNbJyR2rk
nature.com
Nature - A synthetic proteome-scale strategy enables the identification of a diverse range of human proteins that can induce the degradation or stabilization of a target protein in a...
27
145
551