Chaoran Chen
@ChaoranChen_
Followers
361
Following
116
Media
9
Statuses
79
PhD student at @ETH_en and developer of @GenSpectrum
Joined March 2018
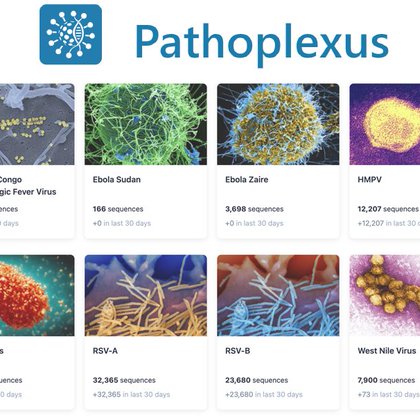
🗓 Pathoplexus turns ONE! 🎂🎉 In the past year we’ve grown from 4 pathogens to a truly global, community-driven platform for transparent, equitable & impactful pathogen sequence sharing. 📰 Read the full update: https://t.co/8tyR0tORdc 1/10
1
11
22
Interested to learn about how to run your own Loculus (the database code behind @pathoplexus)? 🤔🖥️ Sign up for the Loculus Tutorial on Mon 8 Sept at @BC2Conference in Basel, run by @ChaoranChen_ & Anya Parker! 🧬 Register: https://t.co/LI6K36lc4J
#bioinformatics #openScience
0
1
5
📢New job posting!📢 Are you excited by the idea of building global infrastructure to make pathogen sequencing more accessible, interpretable, and equitable? 🧑🏻💻🧬 My group at @SwissTPH has an opening working with ARTIC2, @pathoplexus, & Loculus - read on! 1/5
1
9
15
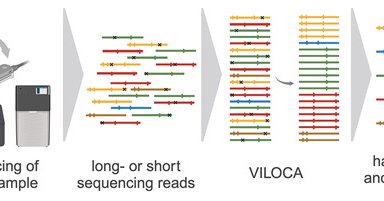
We are excited to share that our new tool VILOCA has been published in NAR G&B: https://t.co/2EfU3U0Ffn! VILOCA is a novel method for mutation calling and local haplotype reconstruction from diverse viral sequencing data, which is also adept for viral surveillance in wastewater.
academic.oup.com
Abstract. RNA viruses exist as large heterogeneous populations within their host. The structure and diversity of virus populations affects disease progress
0
6
9
GenSpectrum just added a mpox dashboard using data from @Pathoplexus – see https://t.co/YFxRxaOePa
0
1
1
Today Pathoplexus announces the inclusion of a new viral pathogen: mpox (previously known as monkeypox). Within the context of the current mpox outbreaks and ongoing health concerns, Pathoplexus aims to support sequence sharing to improve both understanding and response. 1/7
1
15
36
It's wonderful to have initiatives like this to support, recognise, and reward #OpenData work. Congratulations to the other 3 fantastic winners, and to the whole Pathoplexus team, who couldn't all be with us last night! 2/2
0
1
2
We're incredibly honoured to be winners of the @academies_ch @ORData_ch National Prize for Open Research Data 2024! 🎉🎊 "Pathoplexus ensures open access and provides a cutting-edge infrastructure for interactive and programmable data exploration" https://t.co/XXn8vPHsDY 1/2
1
6
19
⭐️And last but not least the fourth Award of the National ORD Prize 2024 goes to ’Pathoplexus’ by Emma Hodcroft, Tanja Stadler, Chaoran Chen, Alexander Taepper, Cornelius Roemer #ordprize24
0
4
11
Elena Dalla Vecchia covers @Pathoplexus in @LancetMicrobe, highlighting that "Pathoplexus offers an advancement in terms of both openness and appropriate accreditation" which hopes to "become a model for similar platforms for other pathogens" https://t.co/KX5w8nZyXR
thelancet.com
On Aug 27, Pathoplexus, a new genomic database for viruses of public health importance, was launched. Its creation—supported by members from ten countries in five continents and developed in consul...
0
8
12
From @PeaseRoland on @bbcworldservice “Pathoplexus helps to protect the people uploading data so that they can share it for surveillance & public health, without feeling like they give up publishing." Listen from 8:26 at https://t.co/YMqxHSYZJ5
#Pathoplexus #OpenScience 1/2
bbc.co.uk
A survey of diseased animals from fur farms has revealed potentially threatening viruses
1
2
6
A great article by @sciencecohen covering @pathoplexus & our values, with quotes from Exec Board member @andersonfbrito_ & the community, Eddie Holmes & @guspalpogeng! And a great excuse to deep-dive into some of the people behind Pathoplexus! 👩🏻🔬👨🏾🔬👨🏿🔬👩🏼🔬 1/9
From @ScienceMagazine: “We believe that building trust through transparency is essential for encouraging broader participation in data sharing,” says Pathoplexus co-founder Anderson Fernandes de Brito @andersonfbrito_
https://t.co/q6Xtfd762t
#Pathoplexus #OpenScience
3
15
53
From @ScienceMagazine: “We believe that building trust through transparency is essential for encouraging broader participation in data sharing,” says Pathoplexus co-founder Anderson Fernandes de Brito @andersonfbrito_
https://t.co/q6Xtfd762t
#Pathoplexus #OpenScience
science.org
Pathoplexus is starting with sequences for Ebola strains and two other risky viruses
0
14
34
It was an amazing journey for us (our research team @ETH_en with the support of contracted software developers from TNG technology) to work with colleagues across the globe from the first idea to what we now call Pathoplexus https://t.co/krMM3TN9hr! We are very proud to 1/2
pathoplexus.org
Pathoplexus is a new, open-source database dedicated to the efficient sharing of human viral pathogen genomic data, fostering global collaboration and public health response.
📢 We're excited to introduce Pathoplexus, a specialized sequence database for human viral pathogens! 🌍 🧬 Launching with 4 viruses, https://t.co/4wGez2nTCr combines modern open-source software with transparent governance to improve global pathogen sequence sharing. 1/14
2
9
40
👋🏻 You can read our full introduction at https://t.co/rqa6Egvq1x, as well as checking out: - ❤️ Our Values ( https://t.co/WYo264UHvx) - 🔓 Open Data Use ( https://t.co/VU73vPmgWz) - 🔒 Restricted Data Use ( https://t.co/Ng9SqwBQks) - 🧑🏻🎓 Our Exec Board ( https://t.co/zCmSzguCTE) 2/14
1
3
13
📢 We're excited to introduce Pathoplexus, a specialized sequence database for human viral pathogens! 🌍 🧬 Launching with 4 viruses, https://t.co/4wGez2nTCr combines modern open-source software with transparent governance to improve global pathogen sequence sharing. 1/14
pathoplexus.org
Pathoplexus is a new, open-source database dedicated to the efficient sharing of human viral pathogen genomic data, fostering global collaboration and public health response.
4
60
105
DECODING GENOMES - from sequences to phylodynamics: We are proud to present our book on how to analyse genomic sequences with an evolutionary perspective. See https://t.co/seRPOVuCyi for a free pdf, Amazon links for print orders, and further information. Great team effort 1/2
8
190
496
Interested in developing dashboards and tools for public health and genomic epidemiology? We are looking for a software engineer to join our GenSpectrum team at @ETH_BSSE in Basel, 🇨🇭CH! https://t.co/2uu0JTj1Fi Please reach out if you have any questions and please share! :)
0
11
7
I am a researcher studying viruses. I have contributed to the sequencing of more than a million genomes deposited in the GISAID database. Last week I found that my access to bulk downloads from GISAID had been cut off without cause or explanation. [1/3]
30
352
1K
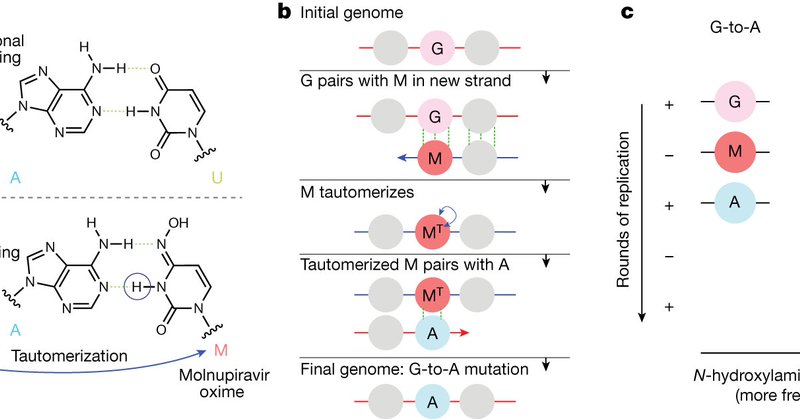
📄 📄 Our molnupiravir work is now out after peer-review! We definitively demonstrate that molnupiravir has resulted in viable SARS-CoV-2 viruses with significant numbers of mutations, in some cases with onwards transmission of mutated viruses.
nature.com
Nature - A specific class of long phylogenetic branches, distinguished by a high proportion of G-to-A and C-to-T mutations, are almost exclusively found in sequences from 2022, after molnupiravir...
84
604
2K