Pathoplexus
@pathoplexus
Followers
374
Following
26
Media
39
Statuses
93
An open-source pathogen sequence database dedicated to equitable sharing, transparent governance, & empowering global public health.
Joined March 2024
12/ 🔗 Read the full update here → https://t.co/ZsNGzDlkO4
#PathogenGenomics #OpenData #OpenScience #PublicHealth #Bioinformatics
pathoplexus.org
Pathoplexus is a new, open-source database dedicated to the efficient sharing of human viral pathogen genomic data, fostering global collaboration and public health response.
0
0
0
11/ 📌 Thank you to YOU - our submitters, users, partners and community - for helping us make Pathoplexus what it is. If you haven’t checked in lately: log in, explore the new features, cite your SeqSets (yes, DOIs matter!), and let us know what you’d like to see next. 🎉💬💻
1
0
0
10/ 🤝 New members: Please join us in welcoming our eight 8 members from Portugal, UK, Uganda (×2), Spain, Ireland (×2), and Bangladesh. Their expertise will help steer Pathoplexus to even greater heights. https://t.co/3OWd4h8RC9
1
0
2
9/ 🏆 Awards & collaborations: We’re excited to share that Loculus was awarded the SIB Remarkable Output 2024 - recognised for enabling efficient & secure sharing of pathogen genomic data: https://t.co/dqWcTYzuV5 Loculus is also joining the Pathogen Data Network!
1
0
2
8/ 🎤 Talks & media: We’ve been on the road! • A 2-part Pathoplexus feature on the MicroBinfie Podcast • Tutorials at BC2, presentations at the IMMEM conference & the PHA4GE Conference & pre-workshop Next up: See us at the Viruses in Silico lecture series (27 Nov 16:00 CET)
1
0
1
7/ 🛠️ Tech improvements (cont.): • Better UI banners for restricted sequences • A new “Tools” tab - which highlights external apps/tools that use Pathoplexus data! If you’ve built one and want yours included, let us know! 👂🏻
1
0
1
6/ 🛠️ Tech improvements: • Support for accented characters in author names • New means2id field for measles sequence upload, linking to the MeaNS2 database • Improved groups pages highlight uploads per pathogen • Multiple choice searches for better filtering!
1
0
1
5/ 🌐 Community & partnerships: We’re proud to now be an institutional member of the International Pathogen Surveillance Network (IPSN) convened by the World Health Organization @WHO - strengthening our global ties and commitment to equitable pathogen data sharing. 🌐🤝
1
1
2
4/ 🧬 On RSV: We updated our classification pipeline — switching from Taxon ID 11250 to 1868215 (Orthopneumovirus) to capture more human RSV sequences. This added ~2,325 RSV-A and ~1,335 RSV-B sequences to our system!
1
1
2
3/ 📊 Data growth (cont.): • 68 new Ebola Zaire virus sequences from DRC (including the first samples from the recent outbreak) • ~1,326 RSV-A & ~1,642 RSV-B sequences from global labs (USA, Australia, Switzerland) 👉 Showing the power of open, shared data.
1
1
2
2/ 📊 Data growth: We now host over 8,900 directly-submitted sequences! Highlights since August include: • ~1,464 new Mpox virus sequences from labs in DRC, Portugal, Ireland, Togo, Canada, Senegal & more • 7 new West Nile virus sequences from Italy & France
1
1
3
1/ 🚀 Big thanks to our community - since August we’ve added thousands of new sequences, rolled out major new features, and welcomed fresh faces into the #Pathoplexus world. Let’s dive into what’s new.👇🏻 Read the full update: https://t.co/ZsNGzDlkO4
2
4
8
We encourage anyone interested in using these data for publications or preprints to reach out to the authors directly. Restricted-Use sharing protects their work and encourages collaborations, while ensuring the sequences are available for public health when needed most. 7/7
0
0
1
Restricted-Use sequences cannot be used for publications or preprints without permission from the data generators. This ensures credit and collaboration, while still supporting urgent response. 6/7
1
0
1
Restricted-Use sequences can be used immediately for public health, outbreak response, and preparedness. This is the main purpose of this option: enabling rapid access when it matters most. 5/7
1
0
1
Since these sequences are likely to attract interest, it’s worth highlighting again what ‘Restricted-Use’ means on Pathoplexus. Data Use Terms for Restricted-Use data: https://t.co/LoASVOGX78 4/7
1
0
1
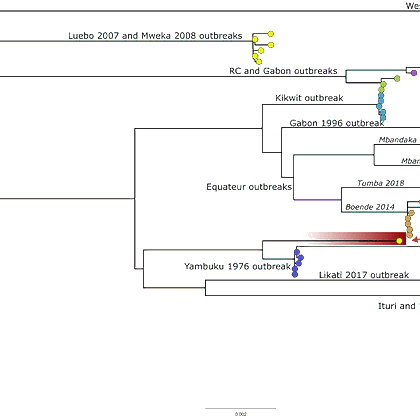
We’re grateful these sequences were shared on Pathoplexus. The authors have chosen to share them as ‘Restricted-Use’ - enabling rapid access for public health, while limiting use in publications/preprints. View the sequences here: https://t.co/Kt3NaH1kUO 3/7
1
0
1
Behind these sequences are the dedicated teams leading the public health response, sampling, testing, & sequencing. With thanks to Placide Mbala-Kingebeni & team at Institute National de Recherche Biomedicale DRC @inrb_kinshasa. See the post for a full author list. 2/7
1
0
1
Amid the concerning news of a new Ebola Zaire outbreak in DRC, teams on the ground have already managed to sample, sequence, and share data. This rapid turnaround is a testament to their commitment and capacity to respond. Read more: https://t.co/z27HV4h96O 1/7
virological.org
Background The Democratic Republic of the Congo (DRC) is currently facing concurrent outbreaks including mpox, cholera and malaria (1-4). The Ministry of Public Health, DRC has declared the 16th...
2
2
16
We are so grateful to our contributors, curators, members & partners worldwide ❤️ Here’s to Year 2 - and an even more connected, equitable future for pathogen genomics! 🎊 Read our whole post here: https://t.co/8tyR0tORdc 10/10
pathoplexus.org
Pathoplexus is a new, open-source database dedicated to the efficient sharing of human viral pathogen genomic data, fostering global collaboration and public health response.
0
0
3