Christoph Hahn
@C__Hahn
Followers
251
Following
276
Media
5
Statuses
1K
Biology, Bioinformatics | husband & father | curious, fascinated, frustrated, exhausted, excited, hungry, surprised ..
Joined October 2014
RT @KeisukeMotone: Our preprint is out! We hacked the @nanopore sequencer to read amino acids and PTMs along protein strands. This opens up….
biorxiv.org
The ability to sequence single protein molecules in their native, full-length form would enable a more comprehensive understanding of proteomic diversity. Current technologies, however, are limited...
0
233
0
RT @jeffnivala: Yesterday we released our preprint showing how a @nanopore array morphs into a protein sequencer. Achieving LONG reads (….
0
35
0
RT @NatureEcoEvo: Evolutionary dynamics of whole-body regeneration across planarian flatworms .
0
55
0
Embracing the command line: my unexpected career in computational biology
nature.com
Nature - A crash course in bioinformatics put Ming Tommy Tang on a different path.
0
1
1
RT @erga_biodiv: Great news from COPO! . Reliably brokering sample #metadata is an essential step of the reference #genome production workf….
0
3
0
RT @rmwaterhouse: Structural #genome annotation with BRAKER & TSEBRA - workshop will take place on the 21st Nov. from 9am to 3pm, registrat….
0
5
0
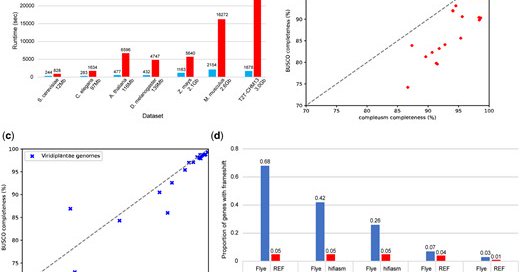
RT @csuhuangneng: Thrilled that our work “compleasm: a faster and more accurate reimplementation of BUSCO” is now published in Bioinformati….
academic.oup.com
AbstractMotivation. Evaluating the gene completeness is critical to measuring the quality of a genome assembly. An incomplete assembly can lead to errors i
0
69
0
RT @steg0s4urus: In a post #AlphaFold world, we can use protein structures in ways we never could before. Can we build phylogenies with th….
0
230
0
Indeed, thanks @philippresl, @EcoEvoGraz, @UniGraz. Check it out - even if you're not interested in #phylogenetics. It's about democratizing complex computational analyses and #reproducibiltiy through #scalability and #portability (#containers). .
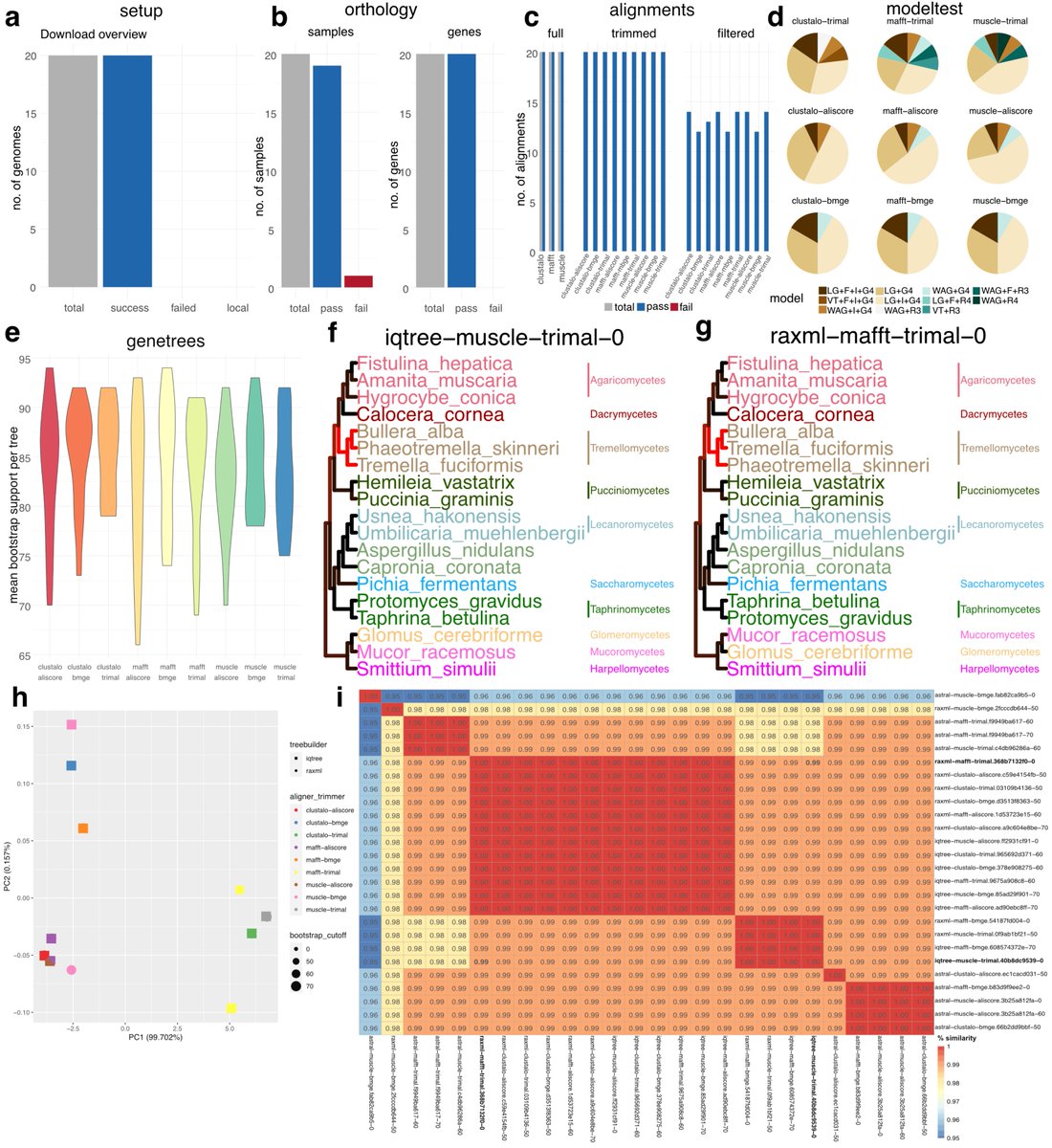
Interested in #phylogenetics using whole #genome/#transcriptome/#proteome data? @C__Hahn and I are very happy to share phylociraptor with you: It facilitates explorative and reproducible phylogenomic analyses of thousands of genes and genomes. Preprint:
0
3
7
RT @philippresl: Interested in #phylogenetics using whole #genome/#transcriptome/#proteome data? @C__Hahn and I are very happy to share phy….
0
123
0
RT @biorxivpreprint: Phylociraptor - A unified computational framework for reproducible phylogenomic inference #bi….
biorxiv.org
Multi-stage phylogenomic analyses require complex computational environments. A unified framework for software- and parameter space exploration, data organisation and -deposition is currently...
0
4
0
RT @biorxiv_bioinfo: Phylociraptor - A unified computational framework for reproducible phylogenomic inference #bi….
0
9
0
RT @MishaKolmogorov: Our CARD pilot paper with @KimberleyBill10 @cornelisblauw @BenedictPaten and many others is online! We establish a wet….
0
16
0
RT @kenbwork: Snakemake has many technical benefits over Nextflow for writing bioinformatics workflows. But there is no platform that supp….
blog.latch.bio
Bioinformatics in Python on the Cloud for Biology
0
30
0
RT @johanneskoester: The small variant calling benchmarking initiative of @NGSCN1 is published. Thanks a lot to all the contributors and @d….
0
11
0
Visual opsin gene expression evolution in the adaptive radiation of cichlid fishes of Lake Tanganyika | Science Advances
science.org
Retinal transcriptomes of Lake Tanganyika cichlids reveal changes in the expression of visual opsin genes associated with ecology.
0
0
0
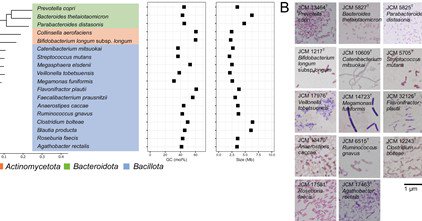
RT @jsantoyo: Assessment of metagenomic workflows using a newly constructed human gut microbiome mock community. #Metagenomics #MicrobiomeM….
academic.oup.com
Abstract. To quantify the biases introduced during human gut microbiome studies, analyzing an artificial mock community as the reference microbiome is indi
0
19
0